Table 3.
Tests of nested models of gene flow for three geographic groups of Batillaria attramentaria in Korea
| Model (Θ) | Log (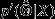 ) ) |
−2 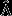
|
P | df |
|---|---|---|---|---|
| m0>1*m0>2 m1>0 m1>2 m2>0 m2>1 m2>3 m3>2 | 2.000 | – | – | – |
| m0>1 = 0 m0>2 m1>0 m1>2 m2>0 m2>1 m2>3 m3>2 | 1.730 | 2.520 | 0.056 | 1 |
| m0>1 m0>2 m1>0 m1>2 m2>0 = 0 m2>1 m2>3 m3>2 | 2.803 | 0.375 | 0.270 | 1 |
| m0>1 m0>2 m1>0 m1>2 m2>0 m2>1 m2>3 = 0 m3>2 | 2.927 | 0.127 | 0.360 | 1 |
| All equal migration rates | 0.538 | 4.904 | 0.972 | 7 |
| All zero migration rates | 2.927 | 0.127 | 0.500 | 8 |
The subscript numbers below migration parameter (m) denote identities of three different geographic populations and an ancestral population (0: YS, 1: SS, 2: JI, and 3: the ancestral population of YS and SS), and the abbreviations of populations are shown in Table1. The symbol “>“between two numbers indicates a direction of gene flow from a recipient population to a donor population in a time-forward manner, as described by Hey and Nielsen (2007). The column heads follow the definition given in detail in the same paper (Hey and Nielsen 2007).
