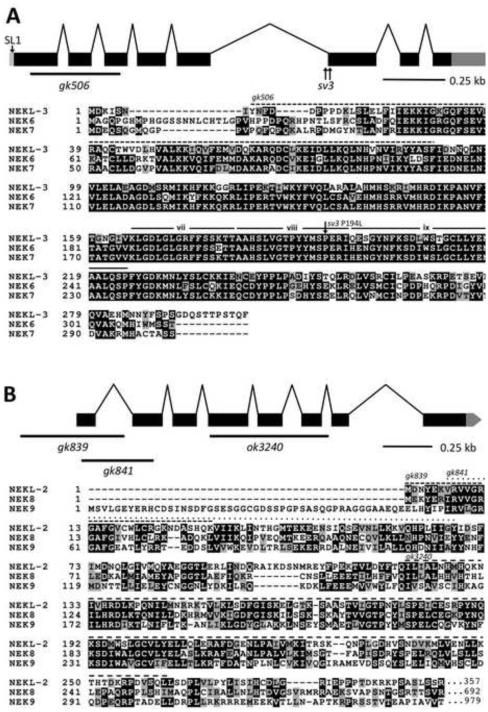
Fig. 2. The structure of nekl-2 and nekl-3 and alignments of their products with human NEKs.
(A) The structure of the nekl-3 locus with the locations of the sv3 alterations and the gk506 deletion is depicted along with an alignment of its predicted amino-acid sequence with human NEK6 and NEK7. The predicted kinase domain of NEKL-3 spans amino acids 23–281; the region deleted by gk506 is indicated by the dashed line. The P194L mutation conferred by sv3 affects a conserved proline in subdomain viii of the kinase domain (subdomains vii, viii and ix are indicated (Hanks and Hunter, 1995)). (B) The structure of the nekl-2 locus with the locations of the gk839, gk841, and ok3240 deletion alleles along with an alignment of its predicted kinase domain (amino acids 4–267) with the kinase domains of human NEK8 and NEK9. The C-terminal halves of both NEK8 and NEK9 have RCC1 repeats (not shown), whereas NEKL-2 extends only for an additional 90 amino acids after the kinase domain (also see supplementary material Fig. S2). In peptide alignments, black boxes with white lettering indicate identical amino acids; gray boxes with black lettering, similar amino acids; white background with black lettering, dissimilar amino acids.