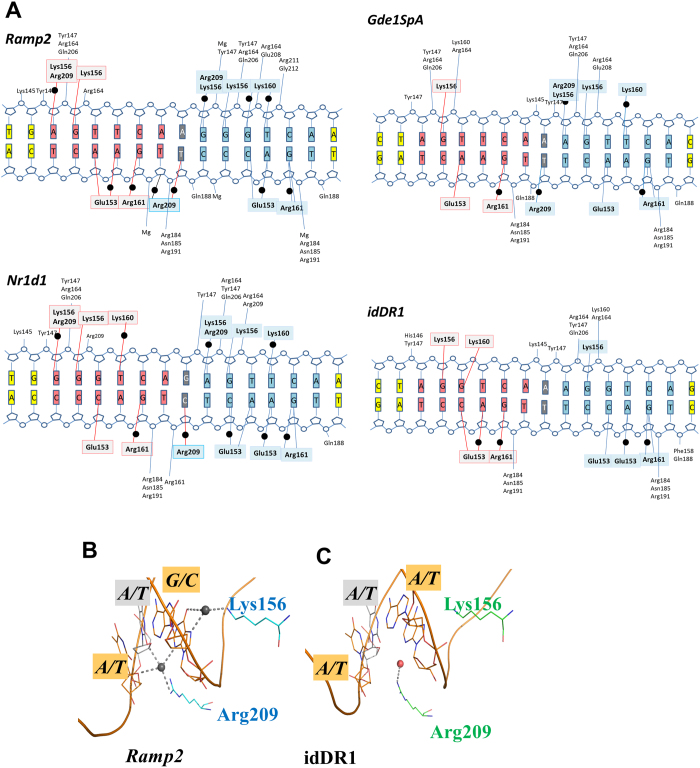
Figure 3. Polarity of the bound RXR homodimers revealed by asymmetric DNA recognition of natural DR1s.
The RXR-DBDs establish unique interactions to recognize natural asymmetric DR1s that are more numerous than in the complex with symmetric idDR1 as revealed by the schematic view of the protein/DNA contacts calculated with NUCPLOT with a 3.9 Å distance cutoff (a). Note that all crystal structures have comparable resolution (between 2 Å and 2.35 Å). For the three natural DR1s studied, the 3′ half-site is more tightly bound with more observed interactions with RXR. Bridging water molecules are shown as black circles. The residues forming hydrogen-bond interactions with the 5′ subunit and 3′subunit are highlighted in light grey and cyan, respectively. The first hexanucleotide is shown in salmon, the second one in blue and the interspacer nucleotide in grey. The gray circles indicate the DNA phosphates and the labeled residues their contacts with RXR. (b-c) Comparison of the interactions around the interspacer nucleotides of the natural Ramp2 and the idealized DR1 complexes. The interspacer is highlighted in grey and the surrounding base pairs in orange. Only the RXR DBD complex with natural DR1 forms specific H-bonds with the interspacer nucleotide. For Ramp2, the downstream DBD extends its interactions upstream of the 3′ half-site to reach the backbone sugar of the last nucleotide of the 5′ site. H-bonds are shown by dashed lines and the water molecules as dark spheres for Ramp2 and red sphere for the idDR1.