Abstract
The β-amyloid precursor protein (APP) has been extensively studied for its role as the precursor of the β-amyloid protein (Aβ) of Alzheimer's disease. However, the normal function of APP remains largely unknown. This article reviews studies on the structure, expression and post-translational processing of APP, as well as studies on the effects of APP in vitro and in vivo. We conclude that the published data provide strong evidence that APP has a trophic function. APP is likely to be involved in neural stem cell development, neuronal survival, neurite outgrowth and neurorepair. However, the mechanisms by which APP exerts its actions remain to be elucidated. The available evidence suggests that APP interacts both intracellularly and extracellularly to regulate various signal transduction mechanisms.
This article reviews studies on the structure, expression and post-translational processing of β-amyloid precursor protein (APP), as well as studies on the effects of APP in vitro and in vivo. We conclude that the published data provide strong evidence that APP has a trophic function. APP is likely to be involved in neural stem cell development, neuronal survival, neurite outgrowth and neurorepair. However, the mechanisms by which APP exerts its actions remain to be elucidated. The available evidence suggests that APP interacts both intracellularly and extracellularly to regulate various signal transduction mechanisms.
Keywords: Alzheimer's disease, amyloid precursor protein, growth factor, heparin, neurotrophic, receptor
The β-amyloid precursor protein (APP) is a type I transmembrane glycoprotein that is expressed in a wide variety of mammalian and non-mammalian cells (Muller-Hill and Beyreuther 1989). APP is the precursor of the β-amyloid protein (Aβ), which is the major protein component of amyloid plaques in the Alzheimer's disease (AD) brain (Masters et al. 1985). Aβ was identified by Glenner and Wong (1984) and the first complete cDNA sequence encoding human APP was cloned in 1987 (Kang et al. 1987). The regulation of APP expression, the mechanisms of APP trafficking, post-translational modification and proteolytic cleavage of APP are now well understood. The production of Aβ from APP, which is generally considered to be a key event in the pathogenesis of AD, has also been well studied. However, despite more than two and a half decades of APP research, the normal function of the protein remains unclear. Circumstantial evidence points towards a number of potential biological roles for APP, but a clearly defined mechanism of action has been elusive. The aim of this article is to examine the putative functions of APP in relation to the expression, post-translational processing and structure of APP.
Expression of APP
APP belongs to a family of evolutionarily and structurally related proteins. The human APP cDNA sequence was first cloned from a brain tissue library (Kang et al. 1987). Subsequently, a number of homologous APP family members were identified in a variety of mammalian and non-mammalian organisms (Muller and Zheng 2012). The APP family in mammals consists of three members: APP, the APP-like protein-1 (APLP1) and the APP-like protein-2 (APLP2) (Wasco et al. 1992, 1993). In humans, the APP gene is located on chromosome 21 (21q21.3), contains 18 exons and extends over a distance of approximately 240 kilobases (Yoshikai et al. 1990) (Fig. 1).
Fig. 1.
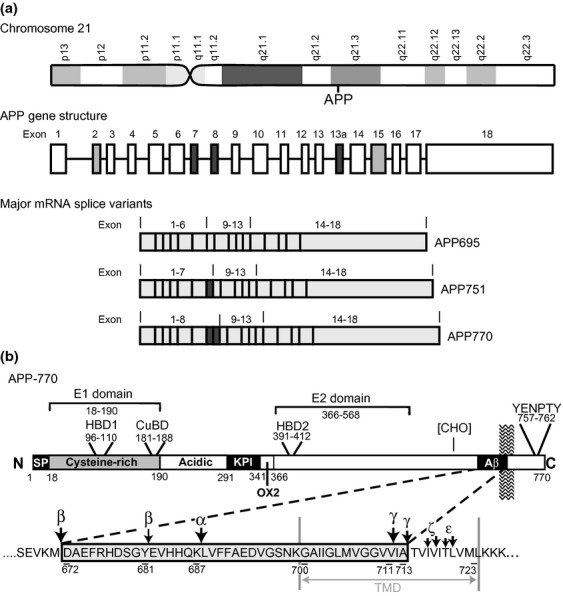
Structure of the amyloid precursor protein (APP) gene, mRNA and protein. (a) The APP gene is located on chromosome 21q21.3. The gene has 18 exons. Differential mRNA splicing of Exons 7,8 (dark grey) can lead to the expression of 695, 751 and 770 amino-acid isoforms. Exons 2 and 15 (light grey) are spliced out in APP639 and L-APP, respectively. (b) Protein structure. APP has an N-terminal signal peptide (SP). The E1 domain has a heparin-binding domain (HBD1), and a copper-binding domain (CuBD); the E2 domain contains a second heparin-binding domain (HBD2). APP751 and APP770 contain a Kunitz protease inhibitor (KPI) and an Ox-2 antigen domain. Between the E2 and Aβ region are two potential N-linked glycosylation sites (CHO). In this region, there is also a potential chondroitin sulphate attachment site that is formed when exon 15 is spliced out. The amino-acid sequence of the Aβ region is shown along with the secretase cleavage sites. The intracellular C-terminal domain contains a YENPTY sorting motif. TMD, transmembrane domain.
The APP promoter sequence indicates that the APP gene belongs to the class of housekeeping genes. The promoter lacks typical TATA and CAAT boxes, but contains consensus sequences for the binding of a number of transcription factors including SP-1, AP-1 and AP-4 sites, a heat shock control element and two Alu-type repetitive sequences (Salbaum et al. 1988; Izumi et al. 1992; Quitschke and Goldgaber 1992). The presence of SP-1, AP-1 and AP-4 sites in the APP promoter, which regulate the expression of proteins associated with cell proliferation and mitosis, as well as cell differentiation, suggests that APP has a function related to cell growth or maturation. Consistent with this idea, the expression of APP or APP-like proteins is increased during development and in association with neurite outgrowth and synaptogenesis (Clarris et al. 1995).
During transcription, differential splicing of APP mRNA can result in a number of APP splice variants (Fig. 1). The major expressed isoforms of APP have 770, 751 or 695 amino acid residues. The APP751 and APP695 isoforms are produced as a result of splicing out of exons 7 and/or 8 (Fig. 1a) (Kang et al. 1987; Tanzi et al. 1988; Weidemann et al. 1989). Some less common splice variants have also been reported, such as L-APP, which lacks exon 15 (Pangalos et al. 1996) and APP639, which lacks exons 2, 7 and 8 (Tang et al. 2003).
APP mRNA is expressed in a wide variety of tissues including the nervous system (brain, spinal cord, retina), immune system (thymus, spleen), muscle (smooth, cardiac and skeletal), kidney, lung, pancreas, prostate gland and thyroid gland (Liu et al. 2008). However, the mRNA splice variants of APP are expressed in different amounts in different cells. APP695 is the predominant neuronal isoform (Kang et al. 1987), but non-neuronal cells express mostly APP770 and APP751 (Rohan de Silva et al. 1997). L-APP is expressed in leucocytes, microglia and astrocytes (Konig et al. 1992). APP639 is expressed widely in foetal tissue, but only in the liver of adults (Tang et al. 2003). The widespread expression, distribution and sequence homology of the APP gene family members suggest that APP plays an important role that is common to many different tissues and organisms.
Gene knock-out (KO) studies can be a powerful method for investigating protein function. APP-KO mice are viable and fertile, indicating that the APP gene alone does not play an essential role in development (Zheng et al. 1995). Similarly, KO of the Drosophila APP homologue (APPL) does not result in a lethal phenotype (Luo et al. 1992). However, APP-KO does result in a number of subtle phenotypic abnormalities. APP-KO mice are slightly smaller, with a reduced weight of 15–20% and reduced brain weight (Zheng et al. 1995; Magara et al. 1999) and APPL-KO in Drosophila results in a behavioural defect (Luo et al. 1992). Importantly, several studies suggest that APP may have a function that is related to the function of other APP family members. Like APP-KO mice, APLP1-KO mice and APLP2-KO mice are both viable and fertile (Heber et al. 2000). Double KO of APLP1 and APP does not produce a lethal phenotype; however, both APP/APLP2 double KO mice and APLP1/APLP2 double KO mice have a postnatal lethal phenotype (Heber et al. 2000). Furthermore, knockout of APL-1, which is the only APP gene in C. elegans, results in a lethal phenotype (Hornsten et al. 2007). Therefore these studies suggest that APP has a function that is likely to be related or overlapping with that of APLP2 in mammals.
Post-translational modification, trafficking and processing of APP
After it has been expressed, the newly translated APP polypeptide can undergo a number of post-translational modifications including glycosylation, sulphation, phosphorylation and palmitoylation (Selkoe 2001; Bhattacharyya et al. 2013). After modification in the Golgi apparatus, APP is trafficked to the cell surface (Koo et al. 1996) before being internalised by clathrin-mediated endocytosis and incorporated into the endosomal-lysosomal system (Yamazaki et al. 1996). Most APP is trafficked from the endosome to the lysosome, where it is degraded (Haass et al. 1992). However, a portion can be returned to the cell surface (Yamazaki et al. 1996).
APP can be post-translationally processed by enzymes termed secretases, which can cleave the protein to produce a number of smaller fragments. The proteolytic processing of APP will not be discussed in detail, as this topic has been well reviewed elsewhere (Haass et al. 2012). APP can initially be cleaved by two proteases, α-secretase or β-secretase (Fig. 1b), to produce the secreted ectodomains sAPPα and sAPPβ. Following APP cleavage by α- or β-secretase, the membrane-associated C-terminal fragments (C83 and C99, respectively) can be cleaved by γ-secretase to yield p3 or Aβ, respectively, and a short C-terminal peptide known as the APP intracellular domain (AICD).
A number of enzymes can act as α-secretases. All of them are members of the A disintegrin and metalloprotease (ADAM) family (Buxbaum et al. 1998; Koike et al. 1999; Lammich et al. 1999). The β-secretase has been identified as a type 1 transmembrane aspartyl protease termed the β-site APP-cleaving enzyme 1 (BACE 1) (Hussain et al. 1999; Sinha et al. 1999; Vassar et al. 1999; Yan et al. 1999; Lin et al. 2000). BACE1 also cleaves APP at position 11 of the Aβ sequence, although the significance of this cleavage is unclear (Fig. 1b) (Liu et al. 2002). γ-Secretase is a transmembrane complex consisting minimally of four protein subunits, presenilin 1 or 2, nicastrin, anterior pharynx-defective phenotype and presenilin enhancer 2 (De Strooper et al. 1998; Yu et al. 2000; Francis et al. 2002; Kimberly et al. 2003). γ-Secretase cleavage is a type of regulated intramembrane proteolysis (RIP), as cleavage occurs in the middle of the transmembrane domain (Lichtenthaler et al. 2011). RIP of APP is thought to occur as a series of cleavages, starting from the C terminal end of the substrate and moving towards the N-terminal region of the transmembrane domain. These cleavage sites have been termed the γ- ε- and ζ- sites (Fig. 1b) (Lichtenthaler et al. 2011).
Although the proteolytic processing of APP by β-secretase can lead to the pathological production of Aβ, β-cleavage is a normal process. Generally, the cleavage of transmembrane proteins by an ADAM or BACE (ectodomain shedding) is commonly involved in the activation of a number of functional pathways. Ectodomain shedding by ADAMs is essential for the release of many cytokines and growth factor ligands, such as epidermal growth factor (EGF) (Blobel 2005). Additionally, ADAMs are involved in ectodomain shedding of growth-factor receptors, such as human epidermal growth factor receptor 2 (Liu et al. 2006) and Notch (Bozkulak and Weinmaster 2009). Ectodomain shedding by BACE is also likely to be required for the proper function of a number of proteins (Klaver et al. 2010). For example, neuregulin is cleaved by BACE1 and ADAM17 to release an ectodomain fragment, which acts in a paracrine manner to stimulate myelination (Fleck et al. 2013). Therefore, cleavage by ADAMs or BACE can potentially facilitate cellular signalling in a variety of ways, either by release of growth factors or by ligand-dependent activation of cellular receptors.
RIP by γ-secretase is also a process involved in the normal function of many proteins. RIP can serve two general functions. First, it can remove the membrane-associated fragment that is produced by ectodomain shedding. Second, it can catalyse the production of intracellular signalling domains (Lichtenthaler et al. 2011). γ-Secretase has over 80 currently known substrates (Haapasalo and Kovacs 2011). Apart from APP, the most well known γ-secretase substrate is the developmental protein Notch, which is activated by γ-secretase cleavage (De Strooper et al. 1999; Struhl and Greenwald 1999). Therefore, it is also possible that γ-secretase cleavage may also be involved in the function of APP.
Structure of APP
Structurally, APP has features of an integral type I transmembrane glycoprotein (Fig. 1b). The structure of APP suggests that it may act as a cell-surface receptor (Kang et al. 1987) or as a growth factor (Rossjohn et al. 1999). The encoded protein contains a large ectodomain, which includes a cysteine-rich globular domain (E1), an acidic domain, a helix-rich domain (E2) and part of the Aβ sequence, which extends into the transmembrane domain (Fig. 1b). The relatively short cytoplasmic domain contains the C-terminus, which has some phosphorylation sites and a YENPTY sorting motif (Fig. 1b). This section will discuss the structure and putative interactions of these domains.
E1 domain and acidic region
The cysteine-rich E1 domain of APP shares little amino-acid sequence similarity to non-APP family members. Cysteine-rich globular domains are found in a number of transmembrane domain proteins including scavenger receptors and hepsin, a cell-surface serine protease (Wu and Parry 2007). The E1 domain is divided into two distinct regions, the heparin-binding domain (HBD) and the copper/metal binding domain (Fig. 2). The HBD is formed of a single α-helix and an anti-parallel β-sheet, with a loop rich in basic residues (95-110) that binds to heparin (Small et al. 1994; Rossjohn et al. 1999) Immediately adjacent to the HBD is a hydrophobic pocket, which could form either a protein-binding site or a dimerisation site (Rossjohn et al. 1999). It has been proposed that this region may dimerise in the presence of heparin (Gralle et al. 2006; Dahms et al. 2010). The size of the putative binding domain at the N-terminus suggests that APP may act as a receptor for a ligand or act as a growth factor (Rossjohn et al. 1999), or may bind to an extracellular matrix component (e.g. proteoglycan) (Small et al. 1994). Adjacent to the HBD is the copper/metal binding domain, which contains a single α-helix and a short β-sheet (Fig. 2). This region can bind several metal ions (Bush et al. 1993). The role of this domain is unclear, but it has been suggested that copper (II) binding and reduction may be a principal function (Multhaup et al. 1996). On the C-terminal side of the E1 domain is an acidic region of unknown significance that is rich in glutamic acid and aspartic acid residues. This region also contains a stretch of seven threonine residues (Kang et al. 1987).
Fig. 2.
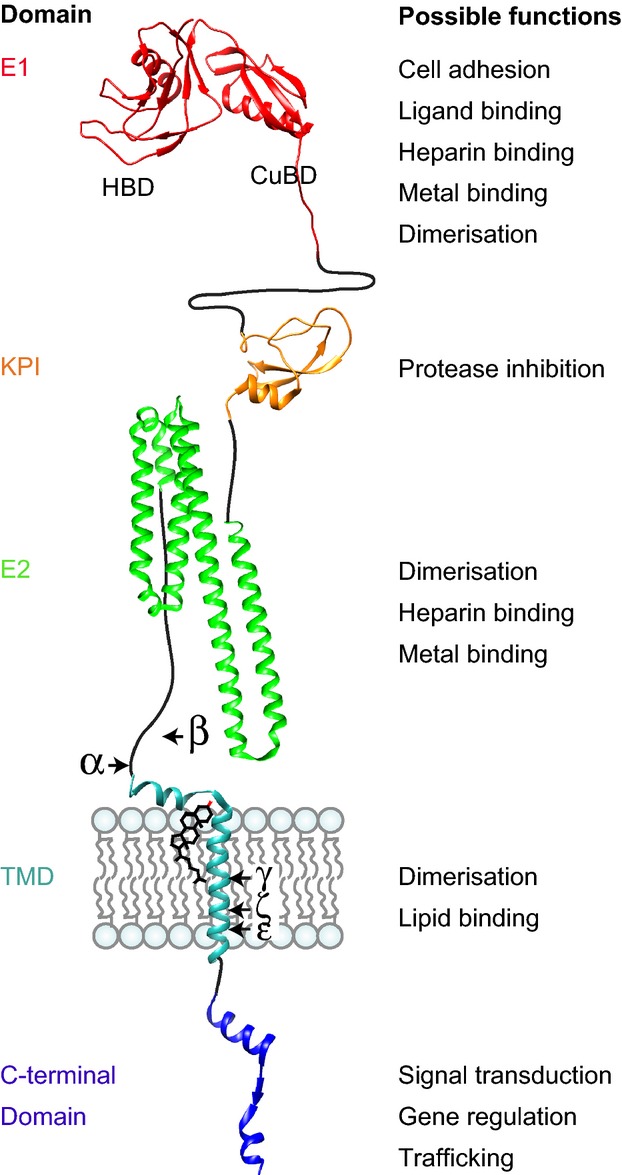
Hypothetical 3-dimensional structure of amyloid precursor protein (APP) based on the following protein data bank files: E1 domain (3KTM), Kunitz protease inhibitor (KPI) domain (1ZJD), E2 domain (3UMK), transmembrane domain (TMD) (2LLM) and intracellular domain (3DXC). A cholesterol molecule is shown in the proposed lipid-binding site in the transmembrane domain. The acidic region and the region between the E2 and Aβ domains are predicated to have little secondary structure.
KPI and Ox-2 antigen domains
Longer isoforms of APP (APP770 and APP751) may contain a Kunitz-type protease inhibitor (KPI) domain and an Ox-2 antigen domain. APP isoforms containing the KPI domain are more commonly expressed in non-neuronal cells (Rohan de Silva et al. 1997), suggesting that they may play a role in glial functions such as in wound repair. Clues to the function of these isoforms comes from studies on blood coagulation. KPI-containing forms of APP (APP751 and APP770) are highly expressed in platelets where they can influence wound repair by regulating blood clotting serine proteases (Van Nostrand et al. 1991b). As serine proteases are also implicated in neuronal cell growth (Wang and Reiser 2003), it is possible that KPI-containing APP isoforms regulate cell growth by inhibiting one or more of these proteases.
The role of the Ox-2 domain in APP770 is less clear. The Ox-2 antigen is a lymphoid and neuronal cell-surface glycoprotein, which has homology to Thy-1 and immunoglobulin light chains (Clark et al. 1985). In APP, the Ox-2 domain is an insert of 19 amino-acid residues that is similar to a region of the Ox-2 antigen. As immunoglobulin loop domains are commonly found in cell-surface receptors and are involved in cell-surface binding and recognition, it seems likely that the Ox-2 domain in APP has a similar function.
E2 domain
The E2 domain is a α-helix rich region (Fig. 2) that can readily dimerize (Xue et al. 2011) and may therefore be involved in APP self-association. The E2 domain has a heparin-binding site (Multhaup 1994; Clarris et al. 1997) as well as a number of putative metal-binding sites that may hold the E2 domain in a rigid conformation (Dahms et al. 2012). The metal-binding site in the E2 domain has been suggested to possess a ferroxidase activity, which may function in cellular iron export through an interaction with ferroportin (Duce et al. 2010). A role in metal homeostasis is supported by the finding that levels of holo-APP can be regulated by iron (Rogers et al. 2002), although more work is needed in this area to support this idea.
Chondroitin sulphate attachment domain
An unusual spliced variant of APP (L-APP) is formed by splicing out exon 15. This creates a consensus sequence for the attachment of chondroitin sulphate (Pangalos et al. 1995). The proteoglycan form of APP (called ‘appican’), which is formed as a consequence of this splicing event, has been found mostly in glia rather than in neurons. Appican contains a chondroitin sulphate E in the repeating disaccharide region and a 4-O-sulphated galactose in the linkage region (Tsuchida et al. 2001). Although the function of appican is unclear, it may be involved in adhesion events (Wu et al. 1997), as chondroitin sulphates are known to inhibit cell attachment and neurite outgrowth (Cui et al. 2013). In addition, appican has been found to bind to certain heparin-binding growth factors such as midkine and pleiotrophin, suggesting that the protein may play some role in the regulation of cell growth (Umehara et al. 2004).
Aβ, transmembrane domain and intracellular domain
The Aβ region on the C-terminal side of the E2 domain lies partly within the ectodomain and partly within the transmembrane domain. A GxxxG sequence motif within the transmembrane domain has been implicated in homodimerisation (Munter et al. 2007) and in cholesterol binding (Barrett et al. 2012; Fig. 2).
From a functional standpoint, the C-terminal cytoplasmic domain of APP is arguably the most interesting region. The structure and possible interactions of this region have been reviewed in detail elsewhere (Kerr and Small 2005; Schettini et al. 2010). The intracellular domain of APP is highly conserved among APP family members and contains a YENPTY sorting motif located between residues 757 and 762 of the APP770 isoform. This motif is involved in the facilitation of clathrin-mediated endocytosis and is present in many tyrosine receptor kinases, non-receptor tyrosine kinases, low-density lipoprotein-receptor related family proteins and integrins (Bonifacino and Traub 2003; Lemmon and Schlessinger 2010). Consistent with this role, many studies have demonstrated that the YENPTY motif in APP is involved in the regulation of its trafficking and endocytosis (Lai et al. 1995; Perez et al. 1999; Ring et al. 2007).
Putative functions of APP
Despite the large number of published studies on APP, there is still no clear consensus on the protein's function. This section aims to summarise the major ideas relating to the function of APP. Coverage of more specific aspects of APP function can be found in other recent reviews (Aydin et al. 2012; Chasseigneaux and Allinquant 2012; Muller and Zheng 2012).
Trophic actions of APP
APP has been reported to influence cell proliferation, differentiation, neurite outgrowth, cell adhesion and synaptogenesis. A number of studies suggest that the extracellular domain can stimulate cellular growth. In vitro, sAPPα has been reported to alter the growth of fibroblasts, keratinocytes, B109 cells, FRTL-5 cells, PC12 cells and neurons (Saitoh et al. 1989; Araki et al. 1991; Milward et al. 1992; Jin et al. 1994; Ninomiya et al. 1994; Pietrzik et al. 1998; Hoffmann et al. 2000; Young-Pearse et al. 2008). Additionally, there are some reports that infusion of sAPPα after traumatic brain injury can improve neuronal survival and recovery (Thornton et al. 2006). Genetic knock-in of sAPPα into APP/APLP2 double KO mice (APPsα-DM mice) rescues the lethal phenotype of the double KO (Weyer et al. 2011), supporting a role for sAPPα in growth. Similarly, knock-in of the extracellular domain fragment of APL-1 from C. elegans rescues the lethal phenotype of the APL-1 KO (Hornsten et al. 2007). Collectively, these studies provide good evidence that APP has a trophic function, and that the extracellular region of APP is involved in this function.
Effects on neural stem cell proliferation and differentiation
As APP is co-ordinately expressed in neuroblasts and neurons at the time of cell proliferation and differentiation (Fukuchi et al. 1992; Masliah et al. 1992; Salbaum and Ruddle 1994; Clarris et al. 1995; Reinhard et al. 2005), this has led to the idea that APP may play a role in the regulation of stem-cell proliferation or differentiation. Indeed, APP is processed in a manner that is very similar to the protein Notch, which regulates neural stem cell differentiation (Ables et al. 2011). Therefore, it is possible that APP may have a similar or related developmental function to that of Notch (Kimberly et al. 2001).
There is strong evidence that APP is able to stimulate the proliferation of neural stem or progenitor cells (NSPCs). For example, sAPPα and sAPPβ can promote the proliferation of NSPCs (Hayashi et al. 1994; Ohsawa et al. 1999; Demars et al. 2011; Baratchi et al. 2012). Hayashi et al. (1994) examined the effect of secreted APP770 on NSPC proliferation and found secreted APP770 had a stronger effect on NSPC proliferation than secreted APP695. A more recent study reported that inhibition of α-secretase reduced NSPC proliferation and that sAPPα was able to rescue this effect (Demars et al. 2011). In another study, sAPPα infused into the ventricles of mice was found to bind to epidermal growth factor receptor (EGFR) expressing stem cells in the subventricular zone (Caille et al. 2004). Both the secretion of EGF and the proliferation of the EGFR-expressing cells were increased by sAPPα infusion (Caille et al. 2004).
To examine the specific contribution of APP to stem cell proliferation, our group examined the ability of NSPCs derived from APP transgenic mice to proliferate. We found that the expression of APP positively correlated with the proliferation of NSPCs (Hu et al. 2013). However, surprisingly, the APP-induced increase in NSPC proliferation was not due to the secretion of sAPPα, but rather to the secretion of cystatin C (Hu et al. 2013). Therefore, APP could potentially influence NSPC proliferation through two different mechanisms, i.e. either via the production of sAPPα or via cystatin C release.
Studies on APP transgenic mice also suggest the possible involvement of APP in NSPC proliferation. Some studies have reported increased NSPC proliferation in APP mice, but have also suggested the effect was due to Aβ (Verret et al. 2007; Kolecki et al. 2008; Sotthibundhu et al. 2009). J20 mice, which overexpress human APP with the Swedish and Indiana familial AD mutations, have a 2-fold increase in the number of proliferating stem cells in the dentate gyrus and subventricular zone at an age of 3 months (Jin et al. 2004; Lopez-Toledano and Shelanski 2007). In contrast, a number of studies have reported decreased NSPC proliferation in APP mice (Haughey et al. 2002; Dong et al. 2004; Donovan et al. 2006; Naumann et al. 2010) or no effect of APP on NSPC proliferation in vivo (Yetman and Jankowsky 2013). As Aβ starts to accumulate in APP mice, the proliferation of neural stem cells decreases (Lopez-Toledano and Shelanski 2007), suggesting that the build-up of Aβ may reduce stem cell proliferation.
APP may also play a role in regulating the differentiation of NSPCs. A study using human embryonic stem cells found that APP overexpression or addition of sAPPα enhanced neuronal differentiation (Freude et al. 2011). We also found that APP-overexpressing NSPCs derived from Tg2576 mice possessed a greater potential to differentiate into neurons, whereas cells derived from APP KO mice exhibited decreased neuronal differentiation (Hu et al. 2013). Another recent study has suggested that sAPPα/β may cause an increase in glial cell differentiation (Baratchi et al. 2012). APP expression is probably not mandatory for the initiation of neuronal differentiation, as embryonic stem cells derived from APP triple KO mice still form neuronal precursors (Bergmans et al. 2010). However, the differentiation of neuronal precursors appears to be delayed in vivo when APP/APLP1 and APLP2 expression is reduced (Shariati et al. 2013).
Effects on neurite outgrowth, synaptogenesis and synaptic plasticity
APP can promote neurite outgrowth in cell culture (Small et al. 1994; Allinquant et al. 1995). Furthermore, APP expression is upregulated rapidly in axons in response to axonal injury, possibly as part of a repair mechanism (Gentleman et al. 1993). One possible mechanism by which APP promotes neurite outgrowth is by regulating cell-substrate adhesion. APP is reported to bind to laminin, collagen type I and heparan sulphate (Kibbey et al. 1993; Beher et al. 1996; Clarris et al. 1997), all of which can influence neurite outgrowth. APP may also promote cell-cell adhesion (Soba et al. 2005). For example, in the presence of heparin, APP can form trans-dimers that could form cell-to-cell contacts (Gralle et al. 2006; Dahms et al. 2010). This trans-dimerisation mode of action has also been proposed as a mechanism for the stabilisation of synapses by APP (Wang et al. 2009). APP may also modulate the activity of other proteins involved in cell adhesion. APP reportedly interacts with several cell-adhesion molecules including integrins, fasciclin II, contactin 4, neuroglia cell adhesion molecule, and transient axonal glycoprotein-1 (Yamazaki et al. 1997; Ashley et al. 2005; Ma et al. 2008; Osterfield et al. 2008). These studies suggest a number of mechanisms by which APP may influence adhesion, although the precise mechanisms still remain obscure.
APP may also be involved in the regulation of synaptogenesis. During development, APP is expressed in both pre- and postsynaptic sites and its level is dramatically increased during the critical period of synaptogenesis (Loffler and Huber 1992; Clarris et al. 1995; Wang et al. 2009). Clarris et al. (1995) found that APP expression was increased in mitral cells of the olfactory bulb at precisely the stage when neurites from olfactory receptor neurons were coming in contact with the mitral cell dendrites. In neurons, a pool of APP is also preferentially found in the post-synapse, suggesting a synaptic role for this protein (Shigematsu et al. 1992).
APP KO mice display a number of neurological deficiencies that may be explained by an effect on synaptogenesis, such as a deficit in grip strength and locomotor activity (Zheng et al. 1995; Ring et al. 2007). APP-KO mice also have a number of deficits that are associated with altered synaptic function, such as hypersensitivity to kainate-induced seizures, alterations in dendritic spine density, and reduced performance in tests of spatial memory (Steinbach et al. 1998; Dawson et al. 1999). APP/APLP2 double KO mice have impaired neuromuscular junction formation, as demonstrated by a reduced number of synaptic vesicles, excessive terminal sprouting, incorrect apposition of pre- and post-synaptic proteins and impaired synaptic transmission (Wang et al. 2005). These synaptic deficits may be responsible for the lethality of the APP/APLP2 double KO (Wang et al. 2005).
A role for APP in regulating synaptic plasticity, learning and memory has also been proposed. APP may alter expression of the GluR2 subunit of the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor, which plays an important role in regulating synaptic calcium permeability (Lee et al. 2010). APP may also affect synaptic calcium by altering cell-surface expression of the NMDA receptor (Cousins et al. 2009; Hoe et al. 2009). As sAPPα is secreted during long-term potentiation (LTP) (Fazeli et al. 1994), it may play a role in the regulation of LTP. However, whether cognitive deficits in APP mice (Chapman et al. 1999) are due to APP-induced disruption of LTP (Weyer et al. 2011) or whether they are due to effects of Aβ, as seems likely (Janus et al. 2000; Morgan et al. 2000), remains to be established. Indeed, some studies suggest that APP can directly increase LTP (Ishida et al. 1997; Seabrook et al. 1999; Taylor et al. 2008).
Function of APP in blood coagulation
The predominant forms of APP in blood contain the KPI domain (Bush et al. 1990; Gardella et al. 1990; Van Nostrand et al. 1991a). These isoforms have been suggested to have a role in the regulation of blood coagulation. In platelets, APP, sAPP and Aβ accumulate in α-granules, which are vesicles that store a variety of clotting factors (Van Nostrand et al. 1991b; Blair and Flaumenhaft 2009). Upon platelet stimulation, APP, sAPP and Aβ are released, along with a number of another components of the coagulation cascade (Bush et al. 1990; Gardella et al. 1990; Smith et al. 1990; Van Nostrand et al. 1990; Smith 1997). The KPI domain of APP is a potent inhibitor of the coagulation factors XIa, IXa and Xa (Smith et al. 1990; Schmaier et al. 1993; Scandura et al. 1997). Factor XIa, for example, is strongly inhibited (Ki = 400 pM) (Smith et al. 1990; Scandura et al. 1997). Notwithstanding the role of the KPI domain, other regions of APP may also participate. For example, inhibition of factor XIa by APP is enhanced in presence of heparin, suggesting an involvement of the heparin-binding regions of APP (Smith et al. 1990). The E1 N-terminal heparin-binding domain of APP is also reported to inhibit the activation of factor XII and to inhibit platelet activation, independently of the KPI domain (Niwano et al. 1995; Henry et al. 1998).
KPI-containing forms of APP can inhibit blood coagulation in vitro, consistent with a role of APP as an inhibitor of coagulation (Schmaier et al. 1993; Annich et al. 1999). Genetic overexpression of APP in mice decreases cerebral thrombosis and also increases the severity of haemorrhage in animal models, whereas KO of APP has the opposite effect (Xu et al. 2005, 2007). The anti-coagulant function of APP is also conserved among APP family members (Xu et al. 2009). Therefore in the circulatory system, other APP family members may also have functions in the clotting cascade.
What is the mechanism of APP signalling?
Despite the many reports of effects of APP on cell proliferation, neurite outgrowth and synaptogenesis, the mechanisms that underlie these effects have not been fully elucidated. The original description of the APP gene noted that the structure of APP resembles a cell-surface receptor (Kang et al. 1987), however a receptor function for APP has not been unequivocally established. A major missing piece of information is the identity of a physiological ligand that activates the APP ‘receptor’. F-spondin has been suggested to be a potential APP ligand (Ho and Sudhof 2004). However, the strongest support for the idea that APP functions as a receptor comes from studies that suggest APP can activate intracellular signal transduction mechanisms.
The C-terminal domain (residues 732-751) has been suggested to be a binding site for G-proteins (Nishimoto et al. 1993). The binding of an extracellular antibody to the N-terminal domain of APP may result in signal transduction by activating the guanosine 5′-triphosphate-binding protein GαO (Okamoto et al. 1995; Murayama et al. 1996). The significance of G-protein coupled APP signalling has yet to be fully elucidated, but studies of the insect APP homologue APPL suggest that an APP-G-protein interaction could be involved in the control of neuronal migration (Ramaker et al. 2013).
Other mechanisms of signal transduction have also been proposed. APP has been suggested to activate gene transcription in a similar manner to Notch, which signals through the γ-secretase-mediated release of the Notch intracellular domain. This domain translocates to the nucleus and activates gene transcription. The AICD fragment of APP has also been reported to translocate to the nucleus (Cupers et al. 2001; Gao and Pimplikar 2001). Normally AICD is prone to degradation (Kimberly et al. 2001). However, AICD can be bound by Fe65, which binds to the YENPTY motif through its phosphotyrosine-binding domain (Fiore et al. 1995). Fe65 binding may help to stabilise AICD (Kimberly et al. 2001). After translocating to the nucleus, the Fe65-bound AICD has been reported to form a transcriptionally active complex in combination with Tat-interactive protein 60 (Tip60), which is a histone acetyltransferase (Cao and Sudhof 2001; Gao and Pimplikar 2001). A number of target genes have been reported for AICD. These genes include KAI1 (Baek et al. 2002), APP, BACE, Tip60 (von Rotz et al. 2004), glycogen synthase kinase-3β (Kim et al. 2003), EGFR (Zhang et al. 2007), p53 (Checler et al. 2007), neprilysin (Belyaev et al. 2009) and low-density lipoprotein-receptor related family proteins (Liu et al. 2007).
Despite the evidence that the AICD may be involved in the regulation of gene transcription, some studies suggest that the role of AICD may not be quite so straightforward. For example, γ-secretase-induced AICD release is not necessary for Tip60 activation (Hass and Yankner 2005). Fe65 has also been reported to signal gene transcription independently of APP (Yang et al. 2006). Additionally, many of the downstream gene targets of the proposed AICD complex have been questioned (Chen and Selkoe 2007; Repetto et al. 2007; Waldron et al. 2008; Aydin et al. 2011) and the mechanism is still unclear (Chen and Selkoe 2007; Waldron et al. 2008).
APP may also exert its physiological effects via the secreted fragments sAPPα or sAPPβ. At present, it is not clear whether secreted APP can activate a specific signal transduction pathway via, for example, a growth factor receptor. Binding studies suggest that there is a high-affinity receptor for secreted APP, which interacts with the E1 domain (Reinhard et al. 2013). However, this receptor has not yet been identified. Some putative APP receptors include β1-integrin, lipoprotein receptor related protein-1, class A scavenger receptor, death receptor 6, p75 neurotrophin receptor and APP itself (Kounnas et al. 1995; Santiago-Garcia et al. 2001; Young-Pearse et al. 2008; Gralle et al. 2009; Nikolaev et al. 2009). However, APP may interact with many other extracellular proteins as well (Bai et al. 2008).
To complicate matters, APP's trophic effects may be mediated via other growth factors. For example, sAPP is able to potentiate the action of nerve growth factor (NGF) (Milward et al. 1992; Wallace et al. 1997; Akar and Wallace 1998). APP can also increase the secretion and expression of cystatin C, which positively modulates the growth of NSPCs. (Hu et al. 2013). APP has been suggested to regulate NGF/tyrosine receptor kinase A signalling, through an intracellular interaction involving the C-terminal YENPTY phosphorylation site (Matrone et al. 2011). Along similar lines, NGF, EGF, and fibroblast growth factor-2 have all been reported to increase the expression of APP (Ohyagi and Tabira 1993; Villa et al. 2001) and NGF, EGF and insulin have been reported to increase the secretion of sAPP (Slack et al. 1995; Solano et al. 2000; Ruiz-Leon and Pascual 2001; Caille et al. 2004). The interplay between these growth factor pathways and APP not only suggest that APP is linked to cellular growth, but also presents a challenge for establishing the direct signalling mechanisms undertaken by APP.
It has also been suggested that the production of Aβ from APP may represent a normal physiological function. However, this suggestion has been controversial. Aβ neither possesses a defined primary structure, nor is it produced as a major pathway of APP processing. Nevertheless, a number of functions for Aβ have been proposed. For example, Aβ has been suggested to be involved in cholesterol transport (Yao and Papadopoulos 2002) and Aβ peptides can increase cell adhesion and neurite outgrowth (Koo et al. 1993). Kamenetz et al. (2003) found that synaptic activity regulated Aβ production and that Aβ, in turn, selectively suppressed excitatory neurotransmission, suggesting that synaptic activity may be regulated by a negative feedback loop involving Aβ secretion. In contrast, a more recent study by Abramov et al. (2009) has suggested that Aβ is a positive endogenous regulator of release probability at hippocampal synapses. The identification of Aβ's normal physiological function (if it has one) is extremely important. As many therapeutic strategies for the treatment of AD aim to prevent Aβ production or increase Aβ clearance from the brain, it is important to ensure that these strategies do not disrupt a normal physiological function.
Summary and conclusions
There is strong evidence that APP plays an important role in cell growth and proliferation. There is also evidence that APP may act as a trophic factor to influence events such as neurite outgrowth and synaptogenesis. As APP is expressed at early stages of nervous system development, APP clearly plays a key role in the growth and maturation of many cells. However, the expression of APP in the mature brain and the up-regulation of APP following traumatic brain injury argue for an important tissue-repair function as well.
Although the role of APP as a growth-regulatory molecule can now be stated with some confidence, the precise mechanism by which APP regulates cell growth is still unclear. The extracellular domain of APP may interact with a cell-surface receptor or a component of the extracellular matrix. The intracellular domain is also undoubtedly important and may interact with a number of cytoplasmic adaptor molecules to facilitate signal transduction or control APP trafficking. However, further research is needed to understand APP's mechanism of action. In particular, future research needs to focus on mechanisms of APP action in which the function of APP is most clearly established. By understanding the mechanism of APP action in well-defined roles (e.g. NSPC proliferation), it may be possible to generalise the findings to understand the mechanisms of APP in relation to other less well-defined roles.
Acknowledgments
This work was funded through grants to DHS from the National Health and Medical Research Council of Australia.
All experiments were conducted in compliance with the ARRIVE guidelines. The authors have no conflict of interest to declare.
Glossary
- Aβ
β-amyloid
- AD
Alzheimer's disease
- ADAM
A disintegrin and metalloprotease
- AICD
APP intracellular domain
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- APLP
APP-like protein
- APP
β-amyloid precursor protein
- BACE
β-site APP-cleaving enzyme 1
- CuBD
copper/metal-binding domain
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- HBD
heparin-binding domain
- KO
knock-out
- KPI
Kunitz protease inhibitor
- LRP
low-density lipoprotein-receptor related family proteins
- LTP
long-term potentiation
- NGF
nerve growth factor
- NSPCs
neural stem or progenitor cells
- RIP
regulated intramembrane proteolysis
- sAPP
secreted β-amyloid precursor protein
- Tip60
Tat-interactive protein 60
- TrkA
tyrosine receptor kinase A
References
- Ables JL, Breunig JJ, Eisch AJ, Rakic P. Not(ch) just development: notch signalling in the adult brain. Nat. Rev. Neurosci. 2011;12:269–283. doi: 10.1038/nrn3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramov E, Dolev I, Fogel H, Ciccotosto GD, Ruff E, Slutsky I. Amyloid-beta as a positive endogenous regulator of release probability at hippocampal synapses. Nat. Neurosci. 2009;12:1567–1576. doi: 10.1038/nn.2433. [DOI] [PubMed] [Google Scholar]
- Akar CA, Wallace WC. Amyloid precursor protein modulates the interaction of nerve growth factor with p75 receptor and potentiates its activation of trkA phosphorylation. Brain Res. Mol. Brain Res. 1998;56:125–132. doi: 10.1016/s0169-328x(98)00037-0. [DOI] [PubMed] [Google Scholar]
- Allinquant B, Hantraye P, Mailleux P, Moya K, Bouillot C, Prochiantz A. Downregulation of amyloid precursor protein inhibits neurite outgrowth in vitro. J. Cell Biol. 1995;128:919–927. doi: 10.1083/jcb.128.5.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annich G, White T, Damm D, Zhao Y, Mahdi F, Meinhardt J, Rebello S, Lucchesi B, Bartlett RH, Schmaier AH. Recombinant Kunitz protease inhibitory domain of the amyloid beta-protein precursor as an anticoagulant in venovenous extracorporeal circulation in rabbits. Thromb. Haemost. 1999;82:1474–1481. [PubMed] [Google Scholar]
- Araki W, Kitaguchi N, Tokushima Y, Ishii K, Aratake H, Shimohama S, Nakamura S, Kimura J. Trophic effect of beta-amyloid precursor protein on cerebral cortical neurons in culture. Biochem. Biophys. Res. Commun. 1991;181:265–271. doi: 10.1016/s0006-291x(05)81412-3. [DOI] [PubMed] [Google Scholar]
- Ashley J, Packard M, Ataman B, Budnik V. Fasciclin II signals new synapse formation through amyloid precursor protein and the scaffolding protein dX11/Mint. J. Neurosci. 2005;25:5943–5955. doi: 10.1523/JNEUROSCI.1144-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin D, Filippov MA, Tschape JA, Gretz N, Prinz M, Eils R, Brors B, Muller UC. Comparative transcriptome profiling of amyloid precursor protein family members in the adult cortex. BMC Genomics. 2011;12:160. doi: 10.1186/1471-2164-12-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin D, Weyer SW, Muller UC. Functions of the APP gene family in the nervous system: insights from mouse models. Exp. Brain Res. 2012;217:423–434. doi: 10.1007/s00221-011-2861-2. [DOI] [PubMed] [Google Scholar]
- Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell. 2002;110:55–67. doi: 10.1016/s0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- Bai Y, Markham K, Chen F, et al. The in vivo brain interactome of the amyloid precursor protein. Mol. Cell. Proteomics. 2008;7:15–34. doi: 10.1074/mcp.M700077-MCP200. [DOI] [PubMed] [Google Scholar]
- Baratchi S, Evans J, Tate WP, Abraham WC, Connor B. Secreted amyloid precursor proteins promote proliferation and glial differentiation of adult hippocampal neural progenitor cells. Hippocampus. 2012;22:1517–1527. doi: 10.1002/hipo.20988. [DOI] [PubMed] [Google Scholar]
- Barrett PJ, Song Y, Van Horn WD, Hustedt EJ, Schafer JM, Hadziselimovic A, Beel AJ, Sanders CR. The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol. Science. 2012;336:1168–1171. doi: 10.1126/science.1219988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beher D, Hesse L, Masters CL, Multhaup G. Regulation of amyloid protein precursor (APP) binding to collagen and mapping of the binding sites on APP and collagen type I. J. Biol. Chem. 1996;271:1613–1620. doi: 10.1074/jbc.271.3.1613. [DOI] [PubMed] [Google Scholar]
- Belyaev ND, Nalivaeva NN, Makova NZ, Turner AJ. Neprilysin gene expression requires binding of the amyloid precursor protein intracellular domain to its promoter: implications for Alzheimer disease. EMBO Rep. 2009;10:94–100. doi: 10.1038/embor.2008.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmans BA, Shariati SA, Habets RL, Verstreken P, Schoonjans L, Muller U, Dotti CG, De Strooper B. Neurons generated from APP/APLP1/APLP2 triple knockout embryonic stem cells behave normally in vitro and in vivo: lack of evidence for a cell autonomous role of the amyloid precursor protein in neuronal differentiation. Stem Cells. 2010;28:399–406. doi: 10.1002/stem.296. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya R, Barren C, Kovacs DM. Palmitoylation of amyloid precursor protein regulates amyloidogenic processing in lipid rafts. J. Neurosci. 2013;33:11169–11183. doi: 10.1523/JNEUROSCI.4704-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair P, Flaumenhaft R. Platelet alpha-granules: basic biology and clinical correlates. Blood Rev. 2009;23:177–189. doi: 10.1016/j.blre.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel CP. ADAMs: key components in EGFR signalling and development. Nat. Rev. Mol. Cell Biol. 2005;6:32–43. doi: 10.1038/nrm1548. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- Bozkulak EC, Weinmaster G. Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling. Mol. Cell. Biol. 2009;29:5679–5695. doi: 10.1128/MCB.00406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush AI, Martins RN, Rumble B, et al. The amyloid precursor protein of Alzheimer's disease is released by human platelets. J. Biol. Chem. 1990;265:15977–15983. [PubMed] [Google Scholar]
- Bush AI, Multhaup G, Moir RD, Williamson TG, Small DH, Rumble B, Pollwein P, Beyreuther K, Masters CL. A novel zinc(II) binding site modulates the function of the beta A4 amyloid protein precursor of Alzheimer's disease. J. Biol. Chem. 1993;268:16109–16112. [PubMed] [Google Scholar]
- Buxbaum JD, Liu KN, Luo Y, Slack JL, Stocking KL, Peschon JJ, Johnson RS, Castner BJ, Cerretti DP, Black RA. Evidence that tumor necrosis factor alpha converting enzyme is involved in regulated alpha-secretase cleavage of the Alzheimer amyloid protein precursor. J. Biol. Chem. 1998;273:27765–27767. doi: 10.1074/jbc.273.43.27765. [DOI] [PubMed] [Google Scholar]
- Caille I, Allinquant B, Dupont E, Bouillot C, Langer A, Muller U, Prochiantz A. Soluble form of amyloid precursor protein regulates proliferation of progenitors in the adult subventricular zone. Development. 2004;131:2173–2181. doi: 10.1242/dev.01103. [DOI] [PubMed] [Google Scholar]
- Cao X, Sudhof TC. A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science. 2001;293:115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- Chapman PF, White GL, Jones MW, et al. Impaired synaptic plasticity and learning in aged amyloid precursor protein transgenic mice. Nat. Neurosci. 1999;2:271–276. doi: 10.1038/6374. [DOI] [PubMed] [Google Scholar]
- Chasseigneaux S, Allinquant B. Functions of Abeta, sAPPalpha and sAPPbeta: similarities and differences. J. Neurochem. 2012;120(Suppl 1):99–108. doi: 10.1111/j.1471-4159.2011.07584.x. [DOI] [PubMed] [Google Scholar]
- Checler F, Sunyach C, Pardossi-Piquard R, Sevalle J, Vincent B, Kawarai T, Girardot N, St George-Hyslop P, da Costa CA. The gamma/epsilon-secretase-derived APP intracellular domain fragments regulate p53. Curr. Alzheimer Res. 2007;4:423–426. doi: 10.2174/156720507781788945. [DOI] [PubMed] [Google Scholar]
- Chen AC, Selkoe DJ. Response to: Pardossi-Piquard et al., “Presenilin-dependent transcriptional control of the abeta-degrading enzyme neprilysin by intracellular domains of betaAPP and APLP”. Neuron 46, 541–554. Neuron. 2007;53:479–483. doi: 10.1016/j.neuron.2007.01.023. [DOI] [PubMed] [Google Scholar]
- Clark MJ, Gagnon J, Williams AF, Barclay AN. MRC OX-2 antigen: a lymphoid/neuronal membrane glycoprotein with a structure like a single immunoglobulin light chain. EMBO J. 1985;4:113–118. doi: 10.1002/j.1460-2075.1985.tb02324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarris HJ, Key B, Beyreuther K, Masters CL, Small DH. Expression of the amyloid protein precursor of Alzheimer's disease in the developing rat olfactory system. Brain Res. Dev. Brain Res. 1995;88:87–95. doi: 10.1016/0165-3806(95)00083-p. [DOI] [PubMed] [Google Scholar]
- Clarris HJ, Cappai R, Heffernan D, Beyreuther K, Masters CL, Small DH. Identification of heparin-binding domains in the amyloid precursor protein of Alzheimer's disease by deletion mutagenesis and peptide mapping. J. Neurochem. 1997;68:1164–1172. doi: 10.1046/j.1471-4159.1997.68031164.x. [DOI] [PubMed] [Google Scholar]
- Cousins SL, Hoey SE, Anne Stephenson F, Perkinton MS. Amyloid precursor protein 695 associates with assembled NR2A- and NR2B-containing NMDA receptors to result in the enhancement of their cell surface delivery. J. Neurochem. 2009;111:1501–1513. doi: 10.1111/j.1471-4159.2009.06424.x. [DOI] [PubMed] [Google Scholar]
- Cui H, Freeman C, Jacobson GA, Small DH. Proteoglycans in the central nervous system: role in development, neural repair, and Alzheimer's disease. IUBMB Life. 2013;65:108–120. doi: 10.1002/iub.1118. [DOI] [PubMed] [Google Scholar]
- Cupers P, Orlans I, Craessaerts K, Annaert W, De Strooper B. The amyloid precursor protein (APP)-cytoplasmic fragment generated by gamma-secretase is rapidly degraded but distributes partially in a nuclear fraction of neurones in culture. J. Neurochem. 2001;78:1168–1178. doi: 10.1046/j.1471-4159.2001.00516.x. [DOI] [PubMed] [Google Scholar]
- Dahms SO, Hoefgen S, Roeser D, Schlott B, Guhrs KH, Than ME. Structure and biochemical analysis of the heparin-induced E1 dimer of the amyloid precursor protein. Proc. Natl Acad. Sci. USA. 2010;107:5381–5386. doi: 10.1073/pnas.0911326107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahms SO, Konnig I, Roeser D, Guhrs KH, Mayer MC, Kaden D, Multhaup G, Than ME. Metal binding dictates conformation and function of the amyloid precursor protein (APP) E2 domain. J. Mol. Biol. 2012;416:438–452. doi: 10.1016/j.jmb.2011.12.057. [DOI] [PubMed] [Google Scholar]
- Dawson GR, Seabrook GR, Zheng H, et al. Age-related cognitive deficits, impaired long-term potentiation and reduction in synaptic marker density in mice lacking the beta-amyloid precursor protein. Neuroscience. 1999;90:1–13. doi: 10.1016/s0306-4522(98)00410-2. [DOI] [PubMed] [Google Scholar]
- De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F. Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature. 1998;391:387–390. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- De Strooper B, Annaert W, Cupers P, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- Demars MP, Bartholomew A, Strakova Z, Lazarov O. Soluble amyloid precursor protein: a novel proliferation factor of adult progenitor cells of ectodermal and mesodermal origin. Stem Cell Res. Ther. 2011;2:36. doi: 10.1186/scrt77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Goico B, Martin M, Csernansky CA, Bertchume A, Csernansky JG. Modulation of hippocampal cell proliferation, memory, and amyloid plaque deposition in APPsw (Tg2576) mutant mice by isolation stress. Neuroscience. 2004;127:601–609. doi: 10.1016/j.neuroscience.2004.05.040. [DOI] [PubMed] [Google Scholar]
- Donovan MH, Yazdani U, Norris RD, Games D, German DC, Eisch AJ. Decreased adult hippocampal neurogenesis in the PDAPP mouse model of Alzheimer's disease. J. Comp. Neurol. 2006;495:70–83. doi: 10.1002/cne.20840. [DOI] [PubMed] [Google Scholar]
- Duce JA, Tsatsanis A, Cater MA, et al. Iron-export ferroxidase activity of beta-amyloid precursor protein is inhibited by zinc in Alzheimer's disease. Cell. 2010;142:857–867. doi: 10.1016/j.cell.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazeli MS, Breen K, Errington ML, Bliss TV. Increase in extracellular NCAM and amyloid precursor protein following induction of long-term potentiation in the dentate gyrus of anaesthetized rats. Neurosci. Lett. 1994;169:77–80. doi: 10.1016/0304-3940(94)90360-3. [DOI] [PubMed] [Google Scholar]
- Fiore F, Zambrano N, Minopoli G, Donini V, Duilio A, Russo T. The regions of the Fe65 protein homologous to the phosphotyrosine interaction/phosphotyrosine binding domain of Shc bind the intracellular domain of the Alzheimer's amyloid precursor protein. J. Biol. Chem. 1995;270:30853–30856. doi: 10.1074/jbc.270.52.30853. [DOI] [PubMed] [Google Scholar]
- Fleck D, van Bebber F, Colombo A, et al. Dual cleavage of neuregulin 1 type III by BACE1 and ADAM17 liberates its EGF-like domain and allows paracrine signaling. J. Neurosci. 2013;33:7856–7869. doi: 10.1523/JNEUROSCI.3372-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R, McGrath G, Zhang J, et al. aph-1 and pen-2 are required for Notch pathway signaling, gamma-secretase cleavage of betaAPP, and presenilin protein accumulation. Dev. Cell. 2002;3:85–97. doi: 10.1016/s1534-5807(02)00189-2. [DOI] [PubMed] [Google Scholar]
- Freude KK, Penjwini M, Davis JL, LaFerla FM, Blurton-Jones M. Soluble amyloid precursor protein induces rapid neural differentiation of human embryonic stem cells. J. Biol. Chem. 2011;286:24264–24274. doi: 10.1074/jbc.M111.227421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi K, Deeb SS, Kamino K, Ogburn CE, Snow AD, Sekiguchi RT, Wight TN, Piussan H, Martin GM. Increased expression of beta-amyloid protein precursor and microtubule-associated protein tau during the differentiation of murine embryonal carcinoma cells. J. Neurochem. 1992;58:1863–1873. doi: 10.1111/j.1471-4159.1992.tb10063.x. [DOI] [PubMed] [Google Scholar]
- Gao Y, Pimplikar SW. The gamma -secretase-cleaved C-terminal fragment of amyloid precursor protein mediates signaling to the nucleus. Proc. Natl Acad. Sci. USA. 2001;98:14979–14984. doi: 10.1073/pnas.261463298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardella JE, Ghiso J, Gorgone GA, Marratta D, Kaplan AP, Frangione B, Gorevic PD. Intact Alzheimer amyloid precursor protein (APP) is present in platelet membranes and is encoded by platelet mRNA. Biochem. Biophys. Res. Commun. 1990;173:1292–1298. doi: 10.1016/s0006-291x(05)80927-1. [DOI] [PubMed] [Google Scholar]
- Gentleman SM, Nash MJ, Sweeting CJ, Graham DI, Roberts GW. Beta-amyloid precursor protein (beta APP) as a marker for axonal injury after head injury. Neurosci. Lett. 1993;160:139–144. doi: 10.1016/0304-3940(93)90398-5. [DOI] [PubMed] [Google Scholar]
- Glenner GG, Wong CW. Alzheimer's disease and Down's syndrome: sharing of a unique cerebrovascular amyloid fibril protein. Biochem. Biophys. Res. Commun. 1984;122:1131–1135. doi: 10.1016/0006-291x(84)91209-9. [DOI] [PubMed] [Google Scholar]
- Gralle M, Oliveira CL, Guerreiro LH, et al. Solution conformation and heparin-induced dimerization of the full-length extracellular domain of the human amyloid precursor protein. J. Mol. Biol. 2006;357:493–508. doi: 10.1016/j.jmb.2005.12.053. [DOI] [PubMed] [Google Scholar]
- Gralle M, Botelho MG, Wouters FS. Neuroprotective secreted amyloid precursor protein acts by disrupting amyloid precursor protein dimers. J. Biol. Chem. 2009;284:15016–15025. doi: 10.1074/jbc.M808755200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haapasalo A, Kovacs DM. The many substrates of presenilin/γ-secretase. J. Alzheimers Dis. 2011;25:3–28. doi: 10.3233/JAD-2011-101065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, Koo EH, Mellon A, Hung AY, Selkoe DJ. Targeting of cell-surface beta-amyloid precursor protein to lysosomes: alternative processing into amyloid-bearing fragments. Nature. 1992;357:500–503. doi: 10.1038/357500a0. [DOI] [PubMed] [Google Scholar]
- Haass C, Kaether C, Thinakaran G, Sisodia S. Trafficking and proteolytic processing of APP. Cold Spring Harb. Perspect. Med. 2012;2:a006270. doi: 10.1101/cshperspect.a006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass MR, Yankner BA. A [gamma]-secretase-independent mechanism of signal transduction by the amyloid precursor protein. J. Biol. Chem. 2005;280:36895–36904. doi: 10.1074/jbc.M502861200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughey NJ, Nath A, Chan SL, Borchard AC, Rao MS, Mattson MP. Disruption of neurogenesis by amyloid beta-peptide, and perturbed neural progenitor cell homeostasis, in models of Alzheimer's disease. J. Neurochem. 2002;83:1509–1524. doi: 10.1046/j.1471-4159.2002.01267.x. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Kashiwagi K, Ohta J, Nakajima M, Kawashima T, Yoshikawa K. Alzheimer amyloid protein precursor enhances proliferation of neural stem cells from fetal rat brain. Biochem. Biophys. Res. Commun. 1994;205:936–943. doi: 10.1006/bbrc.1994.2755. [DOI] [PubMed] [Google Scholar]
- Heber S, Herms J, Gajic V, et al. Mice with combined gene knock-outs reveal essential and partially redundant functions of amyloid precursor protein family members. J. Neurosci. 2000;20:7951–7963. doi: 10.1523/JNEUROSCI.20-21-07951.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry A, Li QX, Galatis D, Hesse L, Multhaup G, Beyreuther K, Masters CL, Cappai R. Inhibition of platelet activation by the Alzheimer's disease amyloid precursor protein. Br. J. Haematol. 1998;103:402–415. doi: 10.1046/j.1365-2141.1998.01005.x. [DOI] [PubMed] [Google Scholar]
- Ho A, Sudhof TC. Binding of F-spondin to amyloid-beta precursor protein: a candidate amyloid-beta precursor protein ligand that modulates amyloid-beta precursor protein cleavage. Proc. Natl Acad. Sci. USA. 2004;101:2548–2553. doi: 10.1073/pnas.0308655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoe HS, Fu Z, Makarova A, et al. The effects of amyloid precursor protein on postsynaptic composition and activity. J. Biol. Chem. 2009;284:8495–8506. doi: 10.1074/jbc.M900141200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann J, Twiesselmann C, Kummer MP, Romagnoli P, Herzog V. A possible role for the Alzheimer amyloid precursor protein in the regulation of epidermal basal cell proliferation. Eur. J. Cell Biol. 2000;79:905–914. doi: 10.1078/0171-9335-00117. [DOI] [PubMed] [Google Scholar]
- Hornsten A, Lieberthal J, Fadia S, et al. APL-1, a Caenorhabditis elegans protein related to the human beta-amyloid precursor protein, is essential for viability. Proc. Natl Acad. Sci. USA. 2007;104:1971–1976. doi: 10.1073/pnas.0603997104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Hung AC, Cui H, Dawkins E, Bolos M, Foa L, Young KM, Small DH. Role of cystatin C in amyloid precursor protein-induced proliferation of neural stem/progenitor cells. J. Biol. Chem. 2013;288:18853–18862. doi: 10.1074/jbc.M112.443671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain I, Powell D, Howlett DR, et al. Identification of a novel aspartic protease (Asp 2) as beta-secretase. Mol. Cell. Neurosci. 1999;14:419–427. doi: 10.1006/mcne.1999.0811. [DOI] [PubMed] [Google Scholar]
- Ishida A, Furukawa K, Keller JN, Mattson MP. Secreted form of beta-amyloid precursor protein shifts the frequency dependency for induction of LTD, and enhances LTP in hippocampal slices. NeuroReport. 1997;8:2133–2137. doi: 10.1097/00001756-199707070-00009. [DOI] [PubMed] [Google Scholar]
- Izumi R, Yamada T, Yoshikai S, Sasaki H, Hattori M, Sakaki Y. Positive and negative regulatory elements for the expression of the Alzheimer's disease amyloid precursor-encoding gene in mouse. Gene. 1992;112:189–195. doi: 10.1016/0378-1119(92)90375-y. [DOI] [PubMed] [Google Scholar]
- Janus C, Pearson J, McLaurin J, et al. A beta peptide immunization reduces behavioural impairment and plaques in a model of Alzheimer's disease. Nature. 2000;408:979–982. doi: 10.1038/35050110. [DOI] [PubMed] [Google Scholar]
- Jin LW, Ninomiya H, Roch JM, Schubert D, Masliah E, Otero DA, Saitoh T. Peptides containing the RERMS sequence of amyloid beta/A4 protein precursor bind cell surface and promote neurite extension. J. Neurosci. 1994;14:5461–5470. doi: 10.1523/JNEUROSCI.14-09-05461.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin K, Galvan V, Xie L, Mao XO, Gorostiza OF, Bredesen DE, Greenberg DA. Enhanced neurogenesis in Alzheimer's disease transgenic (PDGF-APPSw, Ind) mice. Proc. Natl Acad. Sci. USA. 2004;101:13363–13367. doi: 10.1073/pnas.0403678101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamenetz F, Tomita T, Hsieh H, Seabrook G, Borchelt D, Iwatsubo T, Sisodia S, Malinow R. APP processing and synaptic function. Neuron. 2003;37:925–937. doi: 10.1016/s0896-6273(03)00124-7. [DOI] [PubMed] [Google Scholar]
- Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, Grzeschik KH, Multhaup G, Beyreuther K, Muller-Hill B. The precursor of Alzheimer's disease amyloid A4 protein resembles a cell-surface receptor. Nature. 1987;325:733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- Kerr ML, Small DH. Cytoplasmic domain of the beta-amyloid protein precursor of Alzheimer's disease: function, regulation of proteolysis, and implications for drug development. J. Neurosci. Res. 2005;80:151–159. doi: 10.1002/jnr.20408. [DOI] [PubMed] [Google Scholar]
- Kibbey MC, Jucker M, Weeks BS, Neve RL, Van Nostrand WE, Kleinman HK. beta-Amyloid precursor protein binds to the neurite-promoting IKVAV site of laminin. Proc. Natl Acad. Sci. USA. 1993;90:10150–10153. doi: 10.1073/pnas.90.21.10150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Kim EM, Lee JP, et al. C-terminal fragments of amyloid precursor protein exert neurotoxicity by inducing glycogen synthase kinase-3beta expression. FASEB J. 2003;17:1951–1953. doi: 10.1096/fj.03-0106fje. [DOI] [PubMed] [Google Scholar]
- Kimberly WT, Zheng JB, Guenette SY, Selkoe DJ. The intracellular domain of the beta-amyloid precursor protein is stabilized by Fe65 and translocates to the nucleus in a notch-like manner. J. Biol. Chem. 2001;276:40288–40292. doi: 10.1074/jbc.C100447200. [DOI] [PubMed] [Google Scholar]
- Kimberly WT, LaVoie MJ, Ostaszewski BL, Ye W, Wolfe MS, Selkoe DJ. Gamma-secretase is a membrane protein complex comprised of presenilin, nicastrin, Aph-1, and Pen-2. Proc. Natl Acad. Sci. USA. 2003;100:6382–6387. doi: 10.1073/pnas.1037392100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaver DW, Wilce MC, Cui H, Hung AC, Gasperini R, Foa L, Small DH. Is BACE1 a suitable therapeutic target for the treatment of Alzheimer's disease? Current strategies and future directions. Biol. Chem. 2010;391:849–859. doi: 10.1515/BC.2010.089. [DOI] [PubMed] [Google Scholar]
- Koike H, Tomioka S, Sorimachi H, Saido TC, Maruyama K, Okuyama A, Fujisawa-Sehara A, Ohno S, Suzuki K, Ishiura S. Membrane-anchored metalloprotease MDC9 has an alpha-secretase activity responsible for processing the amyloid precursor protein. Biochem J. 1999;343(Pt 2):371–375. [PMC free article] [PubMed] [Google Scholar]
- Kolecki R, Lafauci G, Rubenstein R, Mazur-Kolecka B, Kaczmarski W, Frackowiak J. The effect of amyloidosis-beta and ageing on proliferation of neuronal progenitor cells in APP-transgenic mouse hippocampus and in culture. Acta Neuropathol. 2008;116:419–424. doi: 10.1007/s00401-008-0380-4. [DOI] [PubMed] [Google Scholar]
- Konig G, Monning U, Czech C, Prior R, Banati R, Schreiter-Gasser U, Bauer J, Masters CL, Beyreuther K. Identification and differential expression of a novel alternative splice isoform of the beta A4 amyloid precursor protein (APP) mRNA in leukocytes and brain microglial cells. J. Biol. Chem. 1992;267:10804–10809. [PubMed] [Google Scholar]
- Koo EH, Park L, Selkoe DJ. Amyloid beta-protein as a substrate interacts with extracellular matrix to promote neurite outgrowth. Proc. Natl Acad. Sci. USA. 1993;90:4748–4752. doi: 10.1073/pnas.90.10.4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo EH, Squazzo SL, Selkoe DJ, Koo CH. Trafficking of cell-surface amyloid beta-protein precursor. I. Secretion, endocytosis and recycling as detected by labeled monoclonal antibody. J. Cell Sci. 1996;109:991–998. doi: 10.1242/jcs.109.5.991. [DOI] [PubMed] [Google Scholar]
- Kounnas MZ, Moir RD, Rebeck GW, Bush AI, Argraves WS, Tanzi RE, Hyman BT, Strickland DK. LDL receptor-related protein, a multifunctional ApoE receptor, binds secreted beta-amyloid precursor protein and mediates its degradation. Cell. 1995;82:331–340. doi: 10.1016/0092-8674(95)90320-8. [DOI] [PubMed] [Google Scholar]
- Lai A, Sisodia SS, Trowbridge IS. Characterization of sorting signals in the beta-amyloid precursor protein cytoplasmic domain. J. Biol. Chem. 1995;270:3565–3573. [PubMed] [Google Scholar]
- Lammich S, Kojro E, Postina R, Gilbert S, Pfeiffer R, Jasionowski M, Haass C, Fahrenholz F. Constitutive and regulated alpha-secretase cleavage of Alzheimer's amyloid precursor protein by a disintegrin metalloprotease. Proc. Natl Acad. Sci. USA. 1999;96:3922–3927. doi: 10.1073/pnas.96.7.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KJ, Moussa CE, Lee Y, Sung Y, Howell BW, Turner RS, Pak DT, Hoe HS. Beta amyloid-independent role of amyloid precursor protein in generation and maintenance of dendritic spines. Neuroscience. 2010;169:344–356. doi: 10.1016/j.neuroscience.2010.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler SF, Haass C, Steiner H. Regulated intramembrane proteolysis–lessons from amyloid precursor protein processing. J. Neurochem. 2011;117:779–796. doi: 10.1111/j.1471-4159.2011.07248.x. [DOI] [PubMed] [Google Scholar]
- Lin X, Koelsch G, Wu S, Downs D, Dashti A, Tang J. Human aspartic protease memapsin 2 cleaves the beta-secretase site of beta-amyloid precursor protein. Proc. Natl Acad. Sci. USA. 2000;97:1456–1460. doi: 10.1073/pnas.97.4.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Doms RW, Lee VM. Glu11 site cleavage and N-terminally truncated A beta production upon BACE overexpression. Biochemistry. 2002;41:3128–3136. doi: 10.1021/bi015800g. [DOI] [PubMed] [Google Scholar]
- Liu PC, Liu X, Li Y, et al. Identification of ADAM10 as a major source of HER2 ectodomain sheddase activity in HER2 overexpressing breast cancer cells. Cancer Biol. Ther. 2006;5:657–664. doi: 10.4161/cbt.5.6.2708. [DOI] [PubMed] [Google Scholar]
- Liu Q, Zerbinatti CV, Zhang J, Hoe HS, Wang B, Cole SL, Herz J, Muglia L, Bu G. Amyloid precursor protein regulates brain apolipoprotein E and cholesterol metabolism through lipoprotein receptor LRP1. Neuron. 2007;56:66–78. doi: 10.1016/j.neuron.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yu X, Zack DJ, Zhu H, Qian J. TiGER: a database for tissue-specific gene expression and regulation. BMC Bioinformatics. 2008;9:271. doi: 10.1186/1471-2105-9-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffler J, Huber G. Beta-amyloid precursor protein isoforms in various rat brain regions and during brain development. J. Neurochem. 1992;59:1316–1324. doi: 10.1111/j.1471-4159.1992.tb08443.x. [DOI] [PubMed] [Google Scholar]
- Lopez-Toledano MA, Shelanski ML. Increased neurogenesis in young transgenic mice overexpressing human APP(Sw, Ind) J. Alzheimers Dis. 2007;12:229–240. doi: 10.3233/jad-2007-12304. [DOI] [PubMed] [Google Scholar]
- Luo L, Tully T, White K. Human amyloid precursor protein ameliorates behavioral deficit of flies deleted for Appl gene. Neuron. 1992;9:595–605. doi: 10.1016/0896-6273(92)90024-8. [DOI] [PubMed] [Google Scholar]
- Ma QH, Futagawa T, Yang WL, et al. A TAG1-APP signalling pathway through Fe65 negatively modulates neurogenesis. Nat. Cell Biol. 2008;10:283–294. doi: 10.1038/ncb1690. [DOI] [PubMed] [Google Scholar]
- Magara F, Muller U, Li ZW, Lipp HP, Weissmann C, Stagljar M, Wolfer DP. Genetic background changes the pattern of forebrain commissure defects in transgenic mice underexpressing the beta-amyloid-precursor protein. Proc. Natl Acad. Sci. USA. 1999;96:4656–4661. doi: 10.1073/pnas.96.8.4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masliah E, Mallory M, Ge N, Saitoh T. Amyloid precursor protein is localized in growing neurites of neonatal rat brain. Brain Res. 1992;593:323–328. doi: 10.1016/0006-8993(92)91329-d. [DOI] [PubMed] [Google Scholar]
- Masters CL, Simms G, Weinman NA, Multhaup G, McDonald BL, Beyreuther K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc. Natl Acad. Sci. USA. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrone C, Barbagallo AP, La Rosa LR, Florenzano F, Ciotti MT, Mercanti D, Chao MV, Calissano P, D'Adamio L. APP is phosphorylated by TrkA and regulates NGF/TrkA signaling. J. Neurosci. 2011;31:11756–11761. doi: 10.1523/JNEUROSCI.1960-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milward EA, Papadopoulos R, Fuller SJ, Moir RD, Small D, Beyreuther K, Masters CL. The amyloid protein precursor of Alzheimer's disease is a mediator of the effects of nerve growth factor on neurite outgrowth. Neuron. 1992;9:129–137. doi: 10.1016/0896-6273(92)90228-6. [DOI] [PubMed] [Google Scholar]
- Morgan D, Diamond DM, Gottschall PE, et al. A beta peptide vaccination prevents memory loss in an animal model of Alzheimer's disease. Nature. 2000;408:982–985. doi: 10.1038/35050116. [DOI] [PubMed] [Google Scholar]
- Muller UC, Zheng H. Physiological functions of APP family proteins. Cold Spring Harb. Perspect. Med. 2012;2:a006288. doi: 10.1101/cshperspect.a006288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Hill B, Beyreuther K. Molecular biology of Alzheimer's disease. Annu. Rev. Biochem. 1989;58:287–307. doi: 10.1146/annurev.bi.58.070189.001443. [DOI] [PubMed] [Google Scholar]
- Multhaup G. Identification and regulation of the high affinity binding site of the Alzheimer's disease amyloid protein precursor (APP) to glycosaminoglycans. Biochimie. 1994;76:304–311. doi: 10.1016/0300-9084(94)90163-5. [DOI] [PubMed] [Google Scholar]
- Multhaup G, Schlicksupp A, Hesse L, Beher D, Ruppert T, Masters CL, Beyreuther K. The amyloid precursor protein of Alzheimer's disease in the reduction of copper(II) to copper(I) Science. 1996;271:1406–1409. doi: 10.1126/science.271.5254.1406. [DOI] [PubMed] [Google Scholar]
- Munter LM, Voigt P, Harmeier A, Kaden D, Gottschalk KE, Weise C, Pipkorn R, Schaefer M, Langosch D, Multhaup G. GxxxG motifs within the amyloid precursor protein transmembrane sequence are critical for the etiology of Abeta42. EMBO J. 2007;26:1702–1712. doi: 10.1038/sj.emboj.7601616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama Y, Takeda S, Yonezawa K, Giambarella U, Nishimoto I, Ogata E. Cell surface receptor function of amyloid precursor protein that activates Ser/Thr kinases. Gerontology. 1996;42(Suppl 1):2–11. doi: 10.1159/000213818. [DOI] [PubMed] [Google Scholar]
- Naumann N, Alpar A, Ueberham U, Arendt T, Gartner U. Transgenic expression of human wild-type amyloid precursor protein decreases neurogenesis in the adult hippocampus. Hippocampus. 2010;20:971–979. doi: 10.1002/hipo.20693. [DOI] [PubMed] [Google Scholar]
- Nikolaev A, McLaughlin T, O'Leary DD, Tessier-Lavigne M. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature. 2009;457:981–989. doi: 10.1038/nature07767. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ninomiya H, Roch JM, Jin LW, Saitoh T. Secreted form of amyloid beta/A4 protein precursor (APP) binds to two distinct APP binding sites on rat B103 neuron-like cells through two different domains, but only one site is involved in neuritotropic activity. J. Neurochem. 1994;63:495–500. doi: 10.1046/j.1471-4159.1994.63020495.x. [DOI] [PubMed] [Google Scholar]
- Nishimoto I, Okamoto T, Matsuura Y, Takahashi S, Okamoto T, Murayama Y, Ogata E. Alzheimer amyloid protein precursor complexes with brain GTP-binding protein G(o) Nature. 1993;362:75–79. doi: 10.1038/362075a0. [DOI] [PubMed] [Google Scholar]
- Niwano H, Embury PB, Greenberg BD, Ratnoff OD. Inhibitory action of amyloid precursor protein against human Hageman factor (factor XII) J. Lab. Clin. Med. 1995;125:251–256. [PubMed] [Google Scholar]
- Ohsawa I, Takamura C, Morimoto T, Ishiguro M, Kohsaka S. Amino-terminal region of secreted form of amyloid precursor protein stimulates proliferation of neural stem cells. Eur. J. Neurosci. 1999;11:1907–1913. doi: 10.1046/j.1460-9568.1999.00601.x. [DOI] [PubMed] [Google Scholar]
- Ohyagi Y, Tabira T. Effect of growth factors and cytokines on expression of amyloid beta protein precursor mRNAs in cultured neural cells. Brain Res. Mol. Brain Res. 1993;18:127–132. doi: 10.1016/0169-328x(93)90181-n. [DOI] [PubMed] [Google Scholar]
- Okamoto T, Takeda S, Murayama Y, Ogata E, Nishimoto I. Ligand-dependent G protein coupling function of amyloid transmembrane precursor. J. Biol. Chem. 1995;270:4205–4208. doi: 10.1074/jbc.270.9.4205. [DOI] [PubMed] [Google Scholar]
- Osterfield M, Egelund R, Young LM, Flanagan JG. Interaction of amyloid precursor protein with contactins and NgCAM in the retinotectal system. Development. 2008;135:1189–1199. doi: 10.1242/dev.007401. [DOI] [PubMed] [Google Scholar]
- Pangalos MN, Shioi J, Robakis NK. Expression of the chondroitin sulfate proteoglycans of amyloid precursor (appican) and amyloid precursor-like protein 2. J. Neurochem. 1995;65:762–769. doi: 10.1046/j.1471-4159.1995.65020762.x. [DOI] [PubMed] [Google Scholar]
- Pangalos MN, Shioi J, Efthimiopoulos S, Wu A, Robakis NK. Characterization of appican, the chondroitin sulfate proteoglycan form of the Alzheimer amyloid precursor protein. Neurodegeneration. 1996;5:445–451. doi: 10.1006/neur.1996.0061. [DOI] [PubMed] [Google Scholar]
- Perez RG, Soriano S, Hayes JD, Ostaszewski B, Xia W, Selkoe DJ, Chen X, Stokin GB, Koo EH. Mutagenesis identifies new signals for beta-amyloid precursor protein endocytosis, turnover, and the generation of secreted fragments, including Abeta42. J. Biol. Chem. 1999;274:18851–18856. doi: 10.1074/jbc.274.27.18851. [DOI] [PubMed] [Google Scholar]
- Pietrzik CU, Hoffmann J, Stober K, Chen CY, Bauer C, Otero DA, Roch JM, Herzog V. From differentiation to proliferation: the secretory amyloid precursor protein as a local mediator of growth in thyroid epithelial cells. Proc. Natl Acad. Sci. USA. 1998;95:1770–1775. doi: 10.1073/pnas.95.4.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quitschke WW, Goldgaber D. The amyloid beta-protein precursor promoter. A region essential for transcriptional activity contains a nuclear factor binding domain. J. Biol. Chem. 1992;267:17362–17368. [PubMed] [Google Scholar]
- Ramaker JM, Swanson TL, Copenhaver PF. Amyloid precursor proteins interact with the heterotrimeric G protein Go in the control of neuronal migration. J. Neurosci. 2013;33:10165–10181. doi: 10.1523/JNEUROSCI.1146-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard C, Hebert SS, De Strooper B. The amyloid-beta precursor protein: integrating structure with biological function. EMBO J. 2005;24:3996–4006. doi: 10.1038/sj.emboj.7600860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard C, Borgers M, David G, De Strooper B. Soluble amyloid-beta precursor protein binds its cell surface receptor in a cooperative fashion with glypican and syndecan proteoglycans. J. Cell Sci. 2013;126:4856–4861. doi: 10.1242/jcs.137919. [DOI] [PubMed] [Google Scholar]
- Repetto E, Yoon IS, Zheng H, Kang DE. Presenilin 1 regulates epidermal growth factor receptor turnover and signaling in the endosomal-lysosomal pathway. J. Biol. Chem. 2007;282:31504–31516. doi: 10.1074/jbc.M704273200. [DOI] [PubMed] [Google Scholar]
- Ring S, Weyer SW, Kilian SB, et al. The secreted beta-amyloid precursor protein ectodomain APPs alpha is sufficient to rescue the anatomical, behavioral, and electrophysiological abnormalities of APP-deficient mice. J. Neurosci. 2007;27:7817–7826. doi: 10.1523/JNEUROSCI.1026-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JT, Randall JD, Cahill CM, et al. An iron-responsive element type II in the 5′-untranslated region of the Alzheimer's amyloid precursor protein transcript. J. Biol. Chem. 2002;277:45518–45528. doi: 10.1074/jbc.M207435200. [DOI] [PubMed] [Google Scholar]
- Rohan de Silva HA, Jen A, Wickenden C, Jen LS, Wilkinson SL, Patel AJ. Cell-specific expression of beta-amyloid precursor protein isoform mRNAs and proteins in neurons and astrocytes. Brain Res. Mol. Brain Res. 1997;47:147–156. doi: 10.1016/s0169-328x(97)00045-4. [DOI] [PubMed] [Google Scholar]
- Rossjohn J, Cappai R, Feil SC, et al. Crystal structure of the N-terminal, growth factor-like domain of Alzheimer amyloid precursor protein. Nat. Struct. Biol. 1999;6:327–331. doi: 10.1038/7562. [DOI] [PubMed] [Google Scholar]
- von Rotz RC, Kohli BM, Bosset J, Meier M, Suzuki T, Nitsch RM, Konietzko U. The APP intracellular domain forms nuclear multiprotein complexes and regulates the transcription of its own precursor. J. Cell Sci. 2004;117:4435–4448. doi: 10.1242/jcs.01323. [DOI] [PubMed] [Google Scholar]
- Ruiz-Leon Y, Pascual A. Brain-derived neurotrophic factor stimulates beta-amyloid gene promoter activity by a Ras-dependent/AP-1-independent mechanism in SH-SY5Y neuroblastoma cells. J. Neurochem. 2001;79:278–285. doi: 10.1046/j.1471-4159.2001.00547.x. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Sundsmo M, Roch JM, Kimura N, Cole G, Schubert D, Oltersdorf T, Schenk DB. Secreted form of amyloid beta protein precursor is involved in the growth regulation of fibroblasts. Cell. 1989;58:615–622. doi: 10.1016/0092-8674(89)90096-2. [DOI] [PubMed] [Google Scholar]
- Salbaum JM, Ruddle FH. Embryonic expression pattern of amyloid protein precursor suggests a role in differentiation of specific subsets of neurons. J. Exp. Zool. 1994;269:116–127. doi: 10.1002/jez.1402690205. [DOI] [PubMed] [Google Scholar]
- Salbaum JM, Weidemann A, Lemaire HG, Masters CL, Beyreuther K. The promoter of Alzheimer's disease amyloid A4 precursor gene. EMBO J. 1988;7:2807–2813. doi: 10.1002/j.1460-2075.1988.tb03136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Garcia J, Mas-Oliva J, Innerarity TL, Pitas RE. Secreted forms of the amyloid-beta precursor protein are ligands for the class A scavenger receptor. J. Biol. Chem. 2001;276:30655–30661. doi: 10.1074/jbc.M102879200. [DOI] [PubMed] [Google Scholar]
- Scandura JM, Zhang Y, Van Nostrand WE, Walsh PN. Progress curve analysis of the kinetics with which blood coagulation factor XIa is inhibited by protease nexin-2. Biochemistry. 1997;36:412–420. doi: 10.1021/bi9612576. [DOI] [PubMed] [Google Scholar]
- Schettini G, Govoni S, Racchi M, Rodriguez G. Phosphorylation of APP-CTF-AICD domains and interaction with adaptor proteins: signal transduction and/or transcriptional role–relevance for Alzheimer pathology. J. Neurochem. 2010;115:1299–1308. doi: 10.1111/j.1471-4159.2010.07044.x. [DOI] [PubMed] [Google Scholar]
- Schmaier AH, Dahl LD, Rozemuller AJ, Roos RA, Wagner SL, Chung R, Van Nostrand WE. Protease nexin-2/amyloid beta protein precursor. A tight-binding inhibitor of coagulation factor IXa. J. Clin. Invest. 1993;92:2540–2545. doi: 10.1172/JCI116863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabrook GR, Smith DW, Bowery BJ, et al. Mechanisms contributing to the deficits in hippocampal synaptic plasticity in mice lacking amyloid precursor protein. Neuropharmacology. 1999;38:349–359. doi: 10.1016/s0028-3908(98)00204-4. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Alzheimer's disease: genes, proteins, and therapy. Physiol. Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- Shariati SA, Lau P, Hassan BA, Muller U, Dotti CG, De Strooper B, Gartner A. APLP2 regulates neuronal stem cell differentiation during cortical development. J. Cell Sci. 2013;126:1268–1277. doi: 10.1242/jcs.122440. [DOI] [PubMed] [Google Scholar]
- Shigematsu K, McGeer PL, McGeer EG. Localization of amyloid precursor protein in selective postsynaptic densities of rat cortical neurons. Brain Res. 1992;592:353–357. doi: 10.1016/0006-8993(92)91697-d. [DOI] [PubMed] [Google Scholar]
- Sinha S, Anderson JP, Barbour R, et al. Purification and cloning of amyloid precursor protein beta-secretase from human brain. Nature. 1999;402:537–540. doi: 10.1038/990114. [DOI] [PubMed] [Google Scholar]
- Slack BE, Breu J, Petryniak MA, Srivastava K, Wurtman RJ. Tyrosine phosphorylation-dependent stimulation of amyloid precursor protein secretion by the m3 muscarinic acetylcholine receptor. J. Biol. Chem. 1995;270:8337–8344. doi: 10.1074/jbc.270.14.8337. [DOI] [PubMed] [Google Scholar]
- Small DH, Nurcombe V, Reed G, Clarris H, Moir R, Beyreuther K, Masters CL. A heparin-binding domain in the amyloid protein precursor of Alzheimer's disease is involved in the regulation of neurite outgrowth. J. Neurosci. 1994;14:2117–2127. doi: 10.1523/JNEUROSCI.14-04-02117.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CC. Stimulated release of the beta-amyloid protein of Alzheimer's disease by normal human platelets. Neurosci. Lett. 1997;235:157–159. doi: 10.1016/s0304-3940(97)00738-6. [DOI] [PubMed] [Google Scholar]
- Smith RP, Higuchi DA, Broze GJJ. Platelet coagulation factor XIa-inhibitor, a form of Alzheimer amyloid precursor protein. Science. 1990;248:1126–1128. doi: 10.1126/science.2111585. [DOI] [PubMed] [Google Scholar]
- Soba P, Eggert S, Wagner K, et al. Homo- and heterodimerization of APP family members promotes intercellular adhesion. EMBO J. 2005;24:3624–3634. doi: 10.1038/sj.emboj.7600824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano DC, Sironi M, Bonfini C, Solerte SB, Govoni S, Racchi M. Insulin regulates soluble amyloid precursor protein release via phosphatidyl inositol 3 kinase-dependent pathway. FASEB J. 2000;14:1015–1022. doi: 10.1096/fasebj.14.7.1015. [DOI] [PubMed] [Google Scholar]
- Sotthibundhu A, Li QX, Thangnipon W, Coulson EJ. Abeta(1-42) stimulates adult SVZ neurogenesis through the p75 neurotrophin receptor. Neurobiol. Aging. 2009;30:1975–1985. doi: 10.1016/j.neurobiolaging.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Steinbach JP, Muller U, Leist M, Li ZW, Nicotera P, Aguzzi A. Hypersensitivity to seizures in beta-amyloid precursor protein deficient mice. Cell Death Differ. 1998;5:858–866. doi: 10.1038/sj.cdd.4400391. [DOI] [PubMed] [Google Scholar]
- Struhl G, Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- Tang K, Wang C, Shen C, Sheng S, Ravid R, Jing N. Identification of a novel alternative splicing isoform of human amyloid precursor protein gene, APP639. Eur. J. Neurosci. 2003;18:102–108. doi: 10.1046/j.1460-9568.2003.02731.x. [DOI] [PubMed] [Google Scholar]
- Tanzi RE, McClatchey AI, Lamperti ED, Villa-Komaroff L, Gusella JF, Neve RL. Protease inhibitor domain encoded by an amyloid protein precursor mRNA associated with Alzheimer's disease. Nature. 1988;331:528–530. doi: 10.1038/331528a0. [DOI] [PubMed] [Google Scholar]
- Taylor CJ, Ireland DR, Ballagh I, Bourne K, Marechal NM, Turner PR, Bilkey DK, Tate WP, Abraham WC. Endogenous secreted amyloid precursor protein-alpha regulates hippocampal NMDA receptor function, long-term potentiation and spatial memory. Neurobiol. Dis. 2008;31:250–260. doi: 10.1016/j.nbd.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Thornton E, Vink R, Blumbergs PC, Van Den Heuvel C. Soluble amyloid precursor protein alpha reduces neuronal injury and improves functional outcome following diffuse traumatic brain injury in rats. Brain Res. 2006;1094:38–46. doi: 10.1016/j.brainres.2006.03.107. [DOI] [PubMed] [Google Scholar]
- Tsuchida K, Shioi J, Yamada S, Boghosian G, Wu A, Cai H, Sugahara K, Robakis NK. Appican, the proteoglycan form of the amyloid precursor protein, contains chondroitin sulfate E in the repeating disaccharide region and 4-O-sulfated galactose in the linkage region. J. Biol. Chem. 2001;276:37155–37160. doi: 10.1074/jbc.M105818200. [DOI] [PubMed] [Google Scholar]
- Umehara Y, Yamada S, Nishimura S, Shioi J, Robakis NK, Sugahara K. Chondroitin sulfate of appican, the proteoglycan form of amyloid precursor protein, produced by C6 glioma cells interacts with heparin-binding neuroregulatory factors. FEBS Lett. 2004;557:233–238. doi: 10.1016/s0014-5793(03)01506-0. [DOI] [PubMed] [Google Scholar]
- Van Nostrand WE, Schmaier AH, Farrow JS, Cunningham DD. Protease nexin-II (amyloid beta-protein precursor): a platelet alpha-granule protein. Science. 1990;248:745–748. doi: 10.1126/science.2110384. [DOI] [PubMed] [Google Scholar]
- Van Nostrand WE, Schmaier AH, Farrow JS, Cines DB, Cunningham DD. Protease nexin-2/amyloid beta-protein precursor in blood is a platelet-specific protein. Biochem. Biophys. Res. Commun. 1991a;175:15–21. doi: 10.1016/s0006-291x(05)81193-3. [DOI] [PubMed] [Google Scholar]
- Van Nostrand WE, Schmaier AH, Farrow JS, Cunningham DD. Platelet protease nexin-2/amyloid beta-protein precursor. Possible pathologic and physiologic functions. Ann. N. Y. Acad. Sci. 1991b;640:140–144. doi: 10.1111/j.1749-6632.1991.tb00205.x. [DOI] [PubMed] [Google Scholar]
- Vassar R, Bennett BD, Babu-Khan S, et al. Beta-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- Verret L, Jankowsky JL, Xu GM, Borchelt DR, Rampon C. Alzheimer's-type amyloidosis in transgenic mice impairs survival of newborn neurons derived from adult hippocampal neurogenesis. J. Neurosci. 2007;27:6771–6780. doi: 10.1523/JNEUROSCI.5564-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa A, Latasa MJ, Pascual A. Nerve growth factor modulates the expression and secretion of beta-amyloid precursor protein through different mechanisms in PC12 cells. J. Neurochem. 2001;77:1077–1084. doi: 10.1046/j.1471-4159.2001.00315.x. [DOI] [PubMed] [Google Scholar]
- Waldron E, Isbert S, Kern A, Jaeger S, Martin AM, Hebert SS, Behl C, Weggen S, De Strooper B, Pietrzik CU. Increased AICD generation does not result in increased nuclear translocation or activation of target gene transcription. Exp. Cell Res. 2008;314:2419–2433. doi: 10.1016/j.yexcr.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Wallace WC, Akar CA, Lyons WE. Amyloid precursor protein potentiates the neurotrophic activity of NGF. Brain Res. Mol. Brain Res. 1997;52:201–212. doi: 10.1016/s0169-328x(97)00258-1. [DOI] [PubMed] [Google Scholar]
- Wang H, Reiser G. Thrombin signaling in the brain: the role of protease-activated receptors. Biol. Chem. 2003;384:193–202. doi: 10.1515/BC.2003.021. [DOI] [PubMed] [Google Scholar]
- Wang P, Yang G, Mosier DR, et al. Defective neuromuscular synapses in mice lacking amyloid precursor protein (APP) and APP-Like protein 2. J. Neurosci. 2005;25:1219–1225. doi: 10.1523/JNEUROSCI.4660-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wang B, Yang L, Guo Q, Aithmitti N, Songyang Z, Zheng H. Presynaptic and postsynaptic interaction of the amyloid precursor protein promotes peripheral and central synaptogenesis. J. Neurosci. 2009;29:10788–10801. doi: 10.1523/JNEUROSCI.2132-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasco W, Bupp K, Magendantz M, Gusella JF, Tanzi RE, Solomon F. Identification of a mouse brain cDNA that encodes a protein related to the Alzheimer disease-associated amyloid beta protein precursor. Proc. Natl Acad. Sci. USA. 1992;89:10758–10762. doi: 10.1073/pnas.89.22.10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasco W, Gurubhagavatula S, Paradis MD, Romano DM, Sisodia SS, Hyman BT, Neve RL, Tanzi RE. Isolation and characterization of APLP2 encoding a homologue of the Alzheimer's associated amyloid beta protein precursor. Nat. Genet. 1993;5:95–100. doi: 10.1038/ng0993-95. [DOI] [PubMed] [Google Scholar]
- Weidemann A, Konig G, Bunke D, Fischer P, Salbaum JM, Masters CL, Beyreuther K. Identification, biogenesis, and localization of precursors of Alzheimer's disease A4 amyloid protein. Cell. 1989;57:115–126. doi: 10.1016/0092-8674(89)90177-3. [DOI] [PubMed] [Google Scholar]
- Weyer SW, Klevanski M, Delekate A, et al. APP and APLP2 are essential at PNS and CNS synapses for transmission, spatial learning and LTP. EMBO J. 2011;30:2266–2280. doi: 10.1038/emboj.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Parry G. Hepsin and prostate cancer. Front. Biosci. 2007;12:5052–5059. doi: 10.2741/2447. [DOI] [PubMed] [Google Scholar]
- Wu A, Pangalos MN, Efthimiopoulos S, Shioi J, Robakis NK. Appican expression induces morphological changes in C6 glioma cells and promotes adhesion of neural cells to the extracellular matrix. J. Neurosci. 1997;17:4987–4993. doi: 10.1523/JNEUROSCI.17-13-04987.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Davis J, Miao J, Previti ML, Romanov G, Ziegler K, Van Nostrand WE. Protease nexin-2/amyloid beta-protein precursor limits cerebral thrombosis. Proc. Natl Acad. Sci. USA. 2005;102:18135–18140. doi: 10.1073/pnas.0507798102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Previti ML, Van Nostrand WE. Increased severity of hemorrhage in transgenic mice expressing cerebral protease nexin-2/amyloid beta-protein precursor. Stroke. 2007;38:2598–2601. doi: 10.1161/STROKEAHA.106.480103. [DOI] [PubMed] [Google Scholar]
- Xu F, Previti ML, Nieman MT, Davis J, Schmaier AH, Van Nostrand WE. AbetaPP/APLP2 family of Kunitz serine proteinase inhibitors regulate cerebral thrombosis. J. Neurosci. 2009;29:5666–5670. doi: 10.1523/JNEUROSCI.0095-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Lee S, Wang Y, Ha Y. Crystal structure of the E2 domain of amyloid precursor protein-like protein 1 in complex with sucrose octasulfate. J. Biol. Chem. 2011;286:29748–29757. doi: 10.1074/jbc.M111.219659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Koo EH, Selkoe DJ. Trafficking of cell-surface amyloid beta-protein precursor. II. Endocytosis, recycling and lysosomal targeting detected by immunolocalization. J. Cell Sci. 1996;109:999–1008. doi: 10.1242/jcs.109.5.999. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Koo EH, Selkoe DJ. Cell surface amyloid beta-protein precursor colocalizes with beta 1 integrins at substrate contact sites in neural cells. J. Neurosci. 1997;17:1004–1010. doi: 10.1523/JNEUROSCI.17-03-01004.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R, Bienkowski MJ, Shuck ME, et al. Membrane-anchored aspartyl protease with Alzheimer's disease beta-secretase activity. Nature. 1999;402:533–537. doi: 10.1038/990107. [DOI] [PubMed] [Google Scholar]
- Yang Z, Cool BH, Martin GM, Hu Q. A dominant role for FE65 (APBB1) in nuclear signaling. J. Biol. Chem. 2006;281:4207–4214. doi: 10.1074/jbc.M508445200. [DOI] [PubMed] [Google Scholar]
- Yao ZX, Papadopoulos V. Function of beta-amyloid in cholesterol transport: a lead to neurotoxicity. FASEB J. 2002;16:1677–1679. doi: 10.1096/fj.02-0285fje. [DOI] [PubMed] [Google Scholar]
- Yetman MJ, Jankowsky JL. Wild-type neural progenitors divide and differentiate normally in an amyloid-rich environment. J. Neurosci. 2013;33:17335–17341. doi: 10.1523/JNEUROSCI.1917-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikai S, Sasaki H, Doh-ura K, Furuya H, Sakaki Y. Genomic organization of the human amyloid beta-protein precursor gene. Gene. 1990;87:257–263. doi: 10.1016/0378-1119(90)90310-n. [DOI] [PubMed] [Google Scholar]
- Young-Pearse TL, Chen AC, Chang R, Marquez C, Selkoe DJ. Secreted APP regulates the function of full-length APP in neurite outgrowth through interaction with integrin beta1. Neural Dev. 2008;3:15. doi: 10.1186/1749-8104-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Nishimura M, Arawaka S, et al. Nicastrin modulates presenilin-mediated notch/glp-1 signal transduction and betaAPP processing. Nature. 2000;407:48–54. doi: 10.1038/35024009. [DOI] [PubMed] [Google Scholar]
- Zhang YW, Wang R, Liu Q, Zhang H, Liao FF, Xu H. Presenilin/gamma-secretase-dependent processing of beta-amyloid precursor protein regulates EGF receptor expression. Proc. Natl Acad. Sci. USA. 2007;104:10613–10618. doi: 10.1073/pnas.0703903104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Jiang M, Trumbauer ME, et al. beta-Amyloid precursor protein-deficient mice show reactive gliosis and decreased locomotor activity. Cell. 1995;81:525–531. doi: 10.1016/0092-8674(95)90073-x. [DOI] [PubMed] [Google Scholar]


