Abstract
Among patients newly infected with hepatitis C virus (HCV), only 20–30 % clear the infection spontaneously. In the remaining 70 %, the infection persists causing chronic liver inflammation and disease. It is well established that polymorphisms in host genes, especially in components of the innate immune response, contribute to the phenomenon of spontaneous HCV clearance. RIG-I-like helicases such as MDA-5 are cytoplasmic sensors of viral RNA that are critical for triggering innate immune responses after infection with RNA viruses. We analysed 14 non-synonymous single-nucleotide polymorphisms in RIG-I-like helicase-pathway-genes comparing European patients that spontaneously cleared HCV (n = 285) or had persistent infection (n = 509).
We find that polymorphic haplotypes in the MDA-5 gene IFIH1 encoding histidine at position 843 and threonine at position 946 strongly correlate with the resolution of HCV infection (OR: 16.23 (95 % CI: 3.67 – 71.87); p = 1.1 × 10−6). Overexpression of MDA-5 genetic variants in HEK 293 cells and in a tissue culture model of HCV infection revealed that the histidine 843/threonine 946 variant leads to increased baseline and ligand-induced expression of interferon-induced genes and confers an increased ability to suppress HCV replication.
Conclusion
These data suggest that MDA-5 plays a significant role in the defense against HCV and that polymorphisms in MDA-5 can influence the outcome of HCV infection.
Keywords: Hepatitis C virus infection, liver, RIG-I-like helicase pathway, single nucleotide polymorphism, genetic association, IFIH1, diabetes type I
Introduction
With 170 million people affected worldwide chronic hepatitis C virus (HCV) infection is a major public health problem causing cirrhosis, liver failure and hepatocellular carcinoma. The heterogeneity in HCV clearance within infected individuals – spontaneously as well as in response to treatment with interferon-α and ribavirin – is well documented but poorly understood. Recent genome wide association studies have demonstrated the particularly strong contribution of genetic polymorphisms in innate immunity-related genes of the host to HCV clearance, complementing other known factors such as age at infection, gender, ethnicity and viral factors (1, 2). Among reported genes for cytokines, chemokines, chemokine receptors, MHC molecules and NK cell receptors, single nucleotide polymorphisms (SNPs) in the locus of the type III interferon IL-28B have shown the most consistent and strongest association with spontaneous clearance of HCV and response to therapy with pegylated interferon-α and ribavirin (3–6).
Interferons (IFNs) type I and type III are major innate immune cytokines produced by cells upon viral infection. They trigger transcriptional upregulation of host genes to build a strong line of antiviral defense. In fact, many interferon-stimulated gene products have been identified to contribute to HCV clearance (7, 8). IFNs themselves are produced upon well-studied signaling events, initiated by pattern recognition receptors (PRRs) detecting intracellular pathogens (9). The RIG-I-like helicases (RLHs) retinoic acid-inducible gene-I (RIG-I), melanoma differentiation-associated gene 5 (MDA-5) and laboratory of genetics and physiology gene 2 (Lgp2) form an important family of cytoplasmic PRRs triggering the induction of IFNs upon viral infection. RLHs detect the presence of viruses through the recognition of distinct molecular patterns in viral RNA species present in the cytoplasm of infected cells. The molecular pattern detected by RIG-I consists of RNA containing a 5′-triphosphate moiety in direct proximity to a double-stranded RNA stretch. The pattern detected by MDA-5 is less well defined and requires long double stranded RNA (10–12). Upon binding of their RNA ligands, RIG-I and MDA-5 induce a signaling cascade that involves the mitochondrial antiviral signaling protein (MAVS), the signaling adaptor protein for both receptors. This leads ultimately to the activation of transcription factors and the transcriptional induction of IFNs and interferon-stimulated genes (13, 14). RIG-I and MDA-5 are activated through distinct but partially overlapping groups of viruses.
Based on cell culture models, several studies suggested that the RLH pathway, with an emphasis on RIG-I and MAVS, is critically involved in the recognition of HCV and a major target of immune evasion strategies of HCV (15–18).
These findings led us to examine the effect of genetic polymorphisms in the core RLH pathway genes on HCV infection outcome.
PATIENTS AND METHODS
Study participants
Informed consent was obtained from all patients that participated in the study. The study protocol conforms to relevant ethical guidelines as reflected in approval by the ethics committees of the participating universities and hospitals. The study population (n = 1316) was comprised of two cohorts of patients and a group of control individuals, all of Caucasian origin. The first cohort consisted of 199 patients with chronic hepatitis C (CH) and 76 individuals with spontaneously resolved hepatitis C infection (SRH). The second cohort consisted of 310 patients with chronic hepatitis C and 209 individuals with spontaneously resolved hepatitis C infection. The control group (C) consisted of 520 healthy, unrelated individuals. Members of the first cohort were haemophilic patients recruited from the University Hospital Bonn, Irish patients infected with contaminated anti-D immune globulin (19) and patients recruited by the German Kompetenznetz Hepatitis. The second cohort was recruited from three different liver centers across Germany. Healthy controls were obtained from the blood donor pool of the blood bank in Freiburg. Chronic hepatitis C was defined as detectable anti-HCV antibodies and HCV RNA in the serum for a minimum of 6 months. Individuals designated “spontaneously cleared” had anti-HCV antibodies and undetectable HCV RNA in the serum for a minimum of 100 days. For the control group HCV negativity was defined by the absence of anti-HCV antibodies. Demographic features of the patient and control groups are presented in table 1 and supplementary tables 1.1 and 1.2. For the haplotype correlation analysis of SNP combinations at rs1990760 and rs3747517 in MDA-5 subjects were only included if their chromosomal haplotype could be unambiguously determined. Subjects heterozygous at both gene loci were therefore excluded, since in these cases our genotyping method does not allow to unambiguously assign their alleles to one chromosome. This however is essential for the interpretation that the haplotype codes for amino acids of the same protein. From the combined cohorts, 344 subjects of the group chronically infected with HCV (68%), 173 subjects of the group that cleared HCV spontaneously (67%) and 364 of the healthy controls (70%) could be included.
Table 1.
Study population
| Controls | Patients with spontaneously resolved hepatitis C | Patients with chronic hepatitis C | |
|---|---|---|---|
| Number | 520 | 285 Cohort 1: 76 Cohort 2: 209 |
509 Cohort 1: 199 Cohort 2: 310 |
| Gender | m: 383 (73.7%) f: 137 (26.3%) |
m: 110 (38.6%) f: 175 (61.4%) |
m: 245 (48.1%) f: 264 (51.9%) |
| Origin | Germany: 520 (100%) | Germany: 253 (88.8%) Ireland: 28 (9.8%) Southern Europe: 2 (0.7%) Eastern Europe: 2 (0.7%) |
Germany: 433 (85.1%) Ireland: 48 (9.4%) Eastern Europe: 16 (3.1%) Russia: 10 (2.0%) Southern Europe: 2 (0.4%) |
Genotyping and statistical analysis of genetic data
Genotyping was done by PCR and melting curve analysis in a LightCycler® 480 instrument (Roche Diagnostics, Mannheim, Germany) using fluorescence resonance energy transfer (FRET) probes (TIB MOLBIOL, Berlin, Germany). Data were evaluated using the SPSS 16.0 software (SPSS Inc.-IBM, Armonk, NY) and Fisher’s exact test or Chi square test where appropriate. Haploview software was used to generate linkage disequilibrium plots for the analysed genes. See supplementary methods and tables for additional details.
Cell stimulation and measurement of cytokine production and gene expression
HEK 293 cells were transfected in 96-well plates with the indicated plasmids using lipofectamine 2000 (Invitrogen Life Technologies). 24 hours after transfection, cells were stimulated with poly I:C (polyinosine-polycytidylic acid) (Invivogen, San Diego, California) complexed with lipofectamine 2000. IP-10 and IL-28B were measured in the culture supernatants by ELISA (DuoSet, R&D Systems, Minneapolis, MN and USCN, Life Science Inc., Wuhan, China). Gene expression was analysed by quantitative RT-PCR using gene-specific primers and probes from the Roche universal probe library (Roche Diagnostics, Mannheim, Germany). The relative abundance of target transcripts was normalized to the expression levels of HPRT in experiments with Huh-7.5 cells and to the neomycin phosphotransferase (npt) gene in overexpression experiments in HEK293 cells to egalize potential differences in transfection. See supplementary methods and tables for additional details.
HCV replication assay
HCV replication assays were performed as described previously (17). Briefly, Huh-7.5 cells were plated in 24-well plates and transduced with lentiviral pseudoparticles expressing MDA-5 variants or controls, respectively, and additionally RFP as marker for transduction. At 72h post transduction, cells were infected with HCV Jc1 378-1-YPet (20) at a dose yielding approximately 30–50% infected (GFP+) cells. At 48 h post infection, supernatants were collected and cells were analysed by FACS. Virus titers from the supernatant were determined by TCID50 assay on Huh-7.5 cells, as previously described (21). To determine statistical significance, one-way analysis of variance (ANOVA) was used, followed by Dunnett’s multiple comparison test.
Results
To determine the impact of pattern recognition receptor variants on HCV clearance, we analysed 14 non-synonymous SNPs from the NCBI SNP database in the genes coding for RIG-I, MDA-5, Lgp-2 or their adapter MAVS with an expected minor allele frequency > 1%. When we examined an initial European cohort we detected a significant difference in allele distributions between 76 individuals with spontaneously resolved HCV infection (SRH) and 199 patients with chronic HCV infection (CH) only for SNP rs3747517 in MDA-5. No significant difference in allele distribution was seen for the other SNPs analysed in MDA-5, RIG-I, Lgp2 or MAVS (see table 1 and 2). The frequency of the minor T allele of rs3747517 coding for histidine at AA 843 in MDA-5 was significantly higher in the group of patients that had spontaneously recovered from hepatitis C infection (39.2% SRH versus 27.9% CH, p = 0.01; OR 1.67 (95%-CI: 1.12 – 2.48); see table 2). This was consistent with a significant difference of the genotype distributions for rs3747517 (p = 0.03 see table 3.1). This result could be replicated in a second independently collected European cohort consisting of 209 individuals with spontaneously resolved HCV infection and 310 patients with chronic HCV (p = 0.02; table 3.1). The ability to replicate the result in a second cohort strongly argues against a type I error due to multiple testing as the explanation for the differences in allele distribution.
Table 2.
Allele frequency analysis of non-synonymous, coding SNPs in RIG-I, MDA-5, MAVS and Lgp2 as well as rs12979860 in the IL-28B promoter in individuals with chronic hepatitis C and spontaneously resolved hepatitis C in cohort 1
| SNP ID and alleles | Allele frequency (%) | p-value | OR (95% CI) |
|||
|---|---|---|---|---|---|---|
| Control | CH | SRH | ||||
| RIG-I: R7C (rs10813831) G/A | G A |
73.4 26.6 |
71.6 28.4 |
68.4 31.6 |
0.465 | 0.86 (0.57 – 1.29) |
| D580E (rs17217280) A/T | A T |
84.2 15.8 |
88.8 11.2 |
88.3 11.7 |
0.881 | 1.05 (0.59 – 1.89) |
| S183I (rs11795404) G/T | G T |
100 0 |
100 0 |
100 0 |
– | – |
| T260P (rs35527044) A/C | A C |
100 0 |
100 0 |
100 0 |
– | – |
| I406T (rs951618) T/C | T C |
100 0 |
100 0 |
100 0 |
– | – |
| F789L (rs35253851) C/A | C A |
100 0 |
100 0 |
100 0 |
– | – |
| MDA-5: T946A (rs1990760) T/C | T C |
60.0 40.0 |
62.1 37.9 |
55.3 44.7 |
0.171 | 1.32 (0.52 – 1.10) |
| R843H (rs3747517) C/T |
C T |
72.4 27.6 |
72.1 27.9 |
60.8 39.2 |
0.013 |
1.67 (1.12 – 2.48) |
| H460R (rs10930046) T/C | T C |
98.8 1.2 |
99.2 0.8 |
99.4 0.6 |
0.876 | 1.19 (1.12 – 11.61) |
| I923V (rs35667974) A/G | A G |
97.8 2.2 |
98.8 1.2 |
96.6 3.4 |
0.123 | 2.78 (0.73 – 10.52) |
| Mavs: Q198K (rs7262903) C/A | C A |
85.1 14.9 |
85.5 14.5 |
80.5 19.5 |
0.154 | 0.70 (0.43 – 1.14) |
| S409F (rs7269320) C/T | C T |
84.9 15.1 |
86.1 13.9 |
81.2 18.8 |
0.149 | 1.44 (0.88 – 2.36) |
| Lgp2: Q425R (rs2074158) A/G | A G |
83.3 16.7 |
86.6 13.4 |
88.2 11.8 |
0.673 | 0.87 (0.49 – 1.54) |
| R523Q (rs2074160) G/A | G A |
97.8 2.2 |
97.5 2.5 |
98.1 1.9 |
0.683 | 1.31 (0.36 – 4.83) |
| IL-28B: noncoding (rs12979860) |
T C |
n.d. |
41.8 58.2 |
22.4 77.6 |
2.5 × 10−5 |
2.4 (1.62 – 3.83) |
p-value by x2 test for allele distribution comparing CH versus SRH; CH, chronic hepatitis. SRH, spontaneously resolved hepatitis. OR, odds ratio comparing CH versus SRH. CI, confidence interval. n.d., not determined.
Table 3.1.
Genotype frequencies of MDA-5: R843H (rs3747517), cohort 1 and 2
| Analysed group | CH | SRH | CH | SRH |
|---|---|---|---|---|
|
| ||||
| Cohort 1 | Cohort 2 | |||
| Genotype | Frequency (%) | Frequency (%) | ||
| CC | 105 (52.8%) | 26 (35.1%) | 166 (53.5%) | 107 (51.2%) |
| CT | 77 (38.7%) | 38 (51.4%) | 119 (38.4%) | 69 (33%) |
| TT | 17 (8.5%) | 10 (13.5%) | 25 (8.1%) | 33 (15.8%) |
| p-value | 0.03 | 0.02 | ||
p-value by x2 test for genotype distribution. CH, chronic hepatitis. SRH, spontaneously resolved hepatitis.
Similar to previous studies that identified polymorphisms at the IL-28B locus as the strongest predictors for the outcome of an HCV infection (3–6), we also found a strong difference in the allele distribution in the IL-28B gene SNP rs12979860. As predicted, in our first cohort the C allele was significantly more frequent in patients able to clear the hepatitis C virus spontaneously (77.6% SRH versus 58.2% CH, p = 2.5 × 10−5; OR 2.4 (95%-CI: 1.62 – 3.83); see table 2). To ensure that our results for the polymorphism in MDA-5 are not due to a coincidental over- or underrepresentation of genotypes at the IL-28B locus we analysed allele frequencies at rs3747517 in our first cohort using the genotype at rs12979860 in IL-28B as a covariate. However, histidine at position 843 in MDA-5 still correlated with spontaneous HCV clearance in people carrying the non-protective genotypes CT or TT at the IL-28B locus (36.5% SRH versus 25.2% CH, p = 0.046; OR 1.7). This strongly suggests that the SNP-alleles in MDA-5 have an independent effect that is not explained by the distribution of IL-28B polymorphisms in our cohort.
Combined analysis of the two cohorts for MDA-5 rs3747517 provided further statistical support for an overrepresentation of the T allele at rs3747517 in the group able to clear HCV infection spontaneously (p = 7 × 10−3). In this combined analysis individuals with the homozygous genotype TT at rs3747517 were 1.99-fold (95% CI: 1.27 – 3.13; p = 4 × 10−3) more likely to clear hepatitis C infection than individuals either heterozygous or not carrying the T allele at all (see table 3.2).
Table 3.2.
Allele, genotype and carrier frequencies of MDA-5: R843H (rs3747517), combined cohorts
| Analysed group | Control | CH | SRH | p-value | OR (95% CI) |
|---|---|---|---|---|---|
|
| |||||
| Frequency (%) | |||||
| C | 753 (72.4 %) | 738 (72.5 %) | 373 (65.9 %)( | 7 × 10−3 | 1.36 (1.10 – 1.70) |
| T | 287 (27.6 %) | 280 (27.5 %) | 193 (34.1 %) | ||
| CC | 271 (52.1 %) | 271 (53.2 %) | 133 (47.0 %) | 8 × 10−3 | |
| CT | 211 (40.6 %) | 196 (38.5 %) | 107 (37.8 %) | ||
| TT | 38 (7.3 %) | 42 (8.3 %) | 43 (15.2 %) | ||
| CC or CT | 482 (92.7 %) | 467 (91.7 %) | 240 (84.7 %) | 4 × 10−3 | 1.99 (1.27 – 3.13) |
| TT | 38 (7.3 %) | 42 (8.3 %) | 43 (15.2 %) | ||
| CC | 271 (52.1 %) | 271 (53.2 %) | 133 (46.9 %) | 3 × 10−3 | 2.09 (1.30 – 3.35) |
| TT | 38 (7.3 %) | 42 (8.3 %) | 43 (15.2 %) | ||
p-value by x2 test for allele distribution, genotype distribution, C carrier distribution and CC versus TT distribution comparing CH versus SRH. CH, chronic hepatitis. SRH, spontaneously resolved hepatitis. OR, odds ratio. CI, confidence interval.
When we estimated the extent of linkage disequilibrium (LD) between rs3747517 and the other SNPs in MDA-5 we found strong LD (D′ = 0.765; r2 = 0.362) for the polymorphisms rs3747517 and rs1990760 in MDA-5. Several studies have previously identified a T at MDA-5 rs1990760 leading to threonine at AA 946 as a risk allele for diabetes type I (22, 23). Even though the allele distribution for rs1990760 by itself was not significantly associated with the outcome of HCV infection in either cohort or the combined analysis of both cohorts (see supplementary table 4) the strong LD between the two SNPs prompted us to further examine whether combination of alleles at rs3747517 and rs1990760 might be associated with the outcome of a hepatitis C infection.
The analysis of the combined cohorts identified a haplotype in MDA-5 composed of the minor allele T at rs3747517 and the major allele T at rs1990760 coding for a variant with histidine at position 843 and threonine at position 946, in the further manuscript denoted H843/T946, which was found at reduced frequency in patients with chronic hepatitis C compared to uninfected controls and strongly associated with spontaneous clearance of HCV infection (see figure 1A and supplementary table 5.1). The frequency of the haplotype coding for H843/T946 was significantly higher in the group of patients that spontaneously recovered from hepatitis C infection (6.6 % SRH versus 0.29 % CH; p = 3.4 × 10−10; see figure 1B and supplementary table 5.2). Genotype analysis showed that carriers of this haplotype had a 16-fold higher chance of resolving HCV infection (OR: 16.23 (95 % CI: 3.67 – 71.87); p = 1.1 × 10−6; figure 1C). Notably all homozygous carriers of this haplotype in our study had cleared the infection (Supporting table 5.2). Having seen the strong influence of the haplotype coding for H843/T946, we reevaluated, if the effect we had seen previously by analysing the SNP coding for AA 843 separately, was in fact caused by the haplotype. This was indeed the case: A multivariate analysis of allele frequencies using the genotypes coding for the amino acid at 946 as covariates found no significant effect (p = 0.58) for H843 in subjects negative for T946 while it stayed significant (p = 0.02) in subjects bearing at least one allele coding for T946 and was highly significant in subjects homozygous for T946 (p = 3.1 × 10−9).
Fig 1. The haplotype at rs1990760 and rs3747517 coding for histidine at position 843 and threonine at position 946 in MDA-5 is associated with spontaneous clearance of HCV infection.
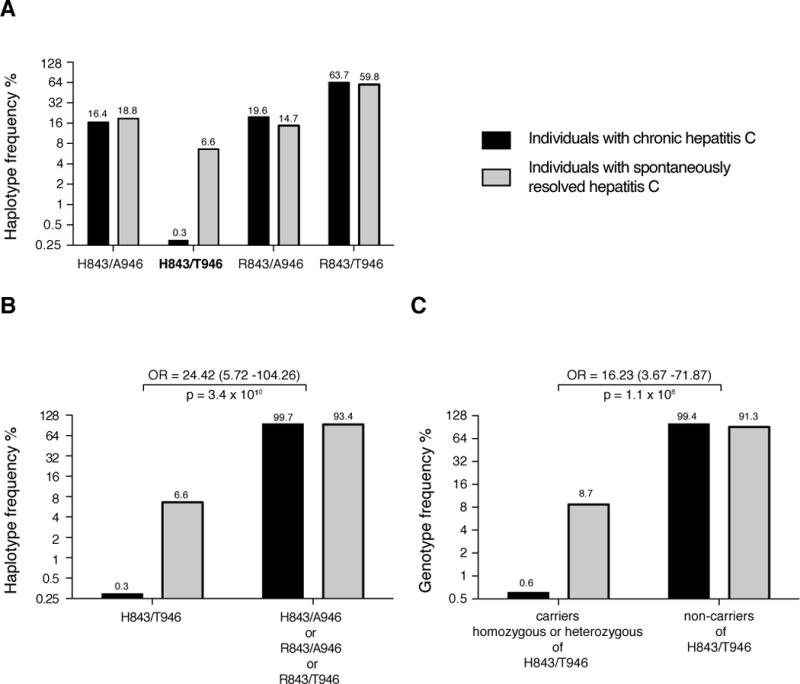
A: The frequency distribution in the combined cohorts 1 and 2 of the four possible MDA-5 haplotypes defined by the two SNPs rs3747517 and rs1990760 is depicted. At rs3747517 the minor allele in the European population (T) codes for histidine at position 843 and the major allele (C) for arginine. At rs1990760 the minor allele (C) codes for alanine at position 946 and the major allele (T) for threonine. The haplotype coding for H843/T946 is suppressed in the group with chronic hepatitis C and associated with clearance of HCV infection. B: The frequency of haplotypes encoding H843/T946 was calculated in the group with chronic hepatitis versus the group with spontaneous resolved hepatitis and compared to the distribution of the combined frequency of the three other possible alleles. C: The frequency of individuals that are carriers, with either heterozygous or homozygous genotypes, of the H843/T946 haplotype versus individuals that do not carry this haplotype was compared in the groups with chronic hepatitis and spontaneous resolved hepatitis respectively. Odds ratios (OR) and 95% confidence intervals are depicted. p-values were calculated using the x2 test.
As the genetic association data indicated that a combination of specific amino acids at position 843 and 946 in MDA-5 might be relevant for the function of the protein and for the immune response against HCV, we performed functional assays to test this hypothesis. Using site-directed mutagenesis we generated constructs of MDA-5 variants containing the four possible amino acid combinations at AAs 843 and 946 encoded by the SNPs at rs3747517 and rs1990760. In addition two previously described (24, 25) naturally occurring, loss-of-function MDA-5 variants, I923V and E627stop, and IRF-1 were included as controls in some experiments. Overexpression of these MDA-5 variants in HEK 293 cells and stimulation with polyinosinic-polycytidylic acid (poly I:C) – a mimic of double-stranded viral RNA and known ligand for MDA-5 – revealed that the H843/T946 variant exhibits enhanced baseline and ligand-induced signaling activity leading to the increased expression of interferon-induced genes (ISG). Upon activation with poly I:C the H843/T946 variant induced higher gene expression of IFN-β, IP-10, ISG15 and ISG56 and the secretion of more IP-10 and IL-28B into the supernatant than R843/T946 – the most common (wild type) variant in the European population – or any other of the tested variants (see figure 2). It is important to note that overexpression of MDA-variants in HEK 293 cells induces a low, but significant, induction of IP-10, IFN-β, ISG15 and ISG56 even in the absence of an added exogenous ligand. The H843/T946 variant showed the highest activity also in this baseline ISG-induction (see figure 2).
Fig 2. The T/T haplotype at rs1990760 and rs3747517 leading to histidine at position 843 and threonine at position 946 encodes a highly active MDA-5 variant.
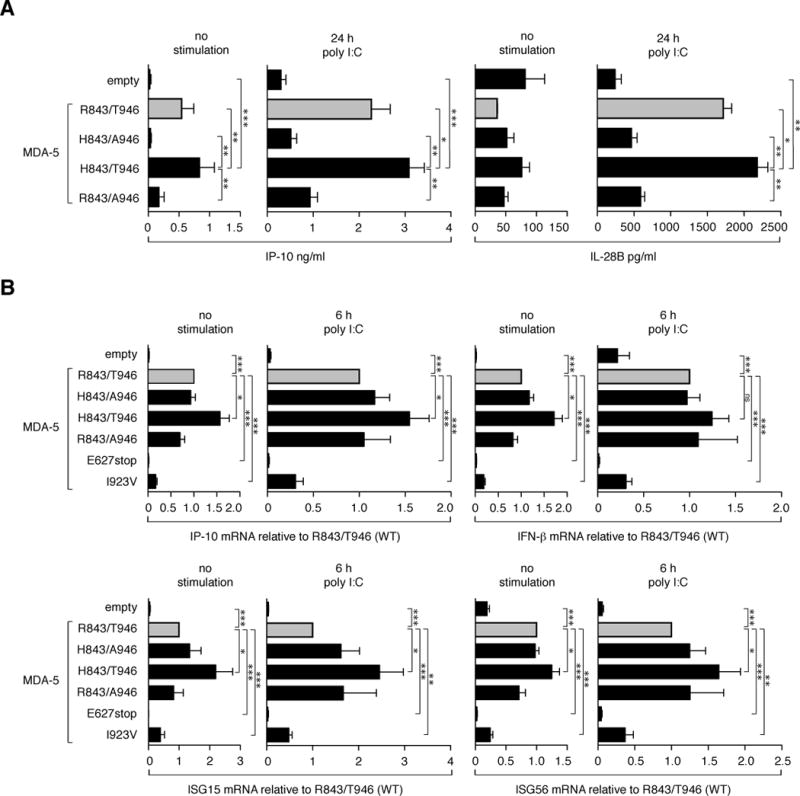
HEK 293 cells were transfected with plasmids coding for the indicated MDA-5 variants (50 ng/well) defined by the four possible amino acid combinations at the amino acid positions 843 and 946 coded by the SNP alleles T/C at rs3747517 and T/C at rs1990760 respectively. R843/T946 the most common allele combination (WT) in the European population is highlighted in grey. An empty vector and in B) two MDA-5 variants, I923V and E627stop, known to be functionally inactive, were used as negative controls. 24 hours after transfection cells were stimulated using poly I:C (125 ng/ml) complexed with lipofectamine for cytoplasmic delivery. A) 24 h after stimulation, production of IP-10 and IL-28B was measured in the supernatants by ELISA. Results are depicted as means + SEM of n = 9 (IP-10) and n =3 (IL-28B). B) Prior to stimulation and 6 h after stimulation RNA was isolated and the expression levels of the indicated genes were analysed by quantitative rt-PCR. Target expression was normalized to the NPT gene and is depicted as fold induction of the WT (R843/T946). Data are depicted as means + SEM of at least n = 5. p-values were determined using Student’s t-test and are depicted as *p<0.05, **p<0.01 and ***p<0.001.
These results encouraged us to examine the function of the MDA-5 variants on HCV replication in a previously established cell culture system of HCV infection (17). Huh-7.5 cells, a RIG-I-defective derivative of Huh-7 human hepatoma cells, were transduced to express the MDA-5 variants and RFP, and subsequently challenged with HCV encoding Ypet as a reporter gene (figure 3A). Viral replication (Ypet-expression) within the RFP-positive population was quantified by FACS analysis (figure 3B). Transduction with an empty vector served as negative control and was set 100% (figure 3C). IRF-1, a potent inhibitor of HCV replication (17), served as positive control. Efficient replication with HCV was seen in the presence of the two functionally inactive MDA-5 variants, I923V and E627stop (see figure 3B). All other MDA-5 variants suppressed HCV replication, albeit with different efficiencies. H843/T946 was significantly more potent than R843/T946 – the most common variant in the European population – or any other tested variant, inhibiting HCV replication by 50%. This potent inhibition by H843/T946 translated to a 100-fold reduction of extracellular HCV titers at 48 h post infection (figure 3D). The increased efficiency is not explained by higher expression of H843/T946, as shown by western blot analyses (see supplementary figure 1). We further determined whether the enhanced virus inhibition by H843/T946 correlated with enhanced ISG expression upon HCV infection. Of note, no ISG stimulation by HCV could be detected without overexpression of a functionally active MDA-5 (figure 3E). For IP-10, ISG15 and ISG56, H843/T946 showed induction even in the absence of HCV, albeit not statistically significant. Furthermore, and in concordance to our previous experiments in HEK 293 cells and poly I:C stimulation (figure 2), we found H843/T946 to be most potent in inducing ISG expression upon infection.
Fig 3. The MDA-5 variant with histidine at position 843 and threonine at position 946 has an increased ability to inhibit HCV replication.
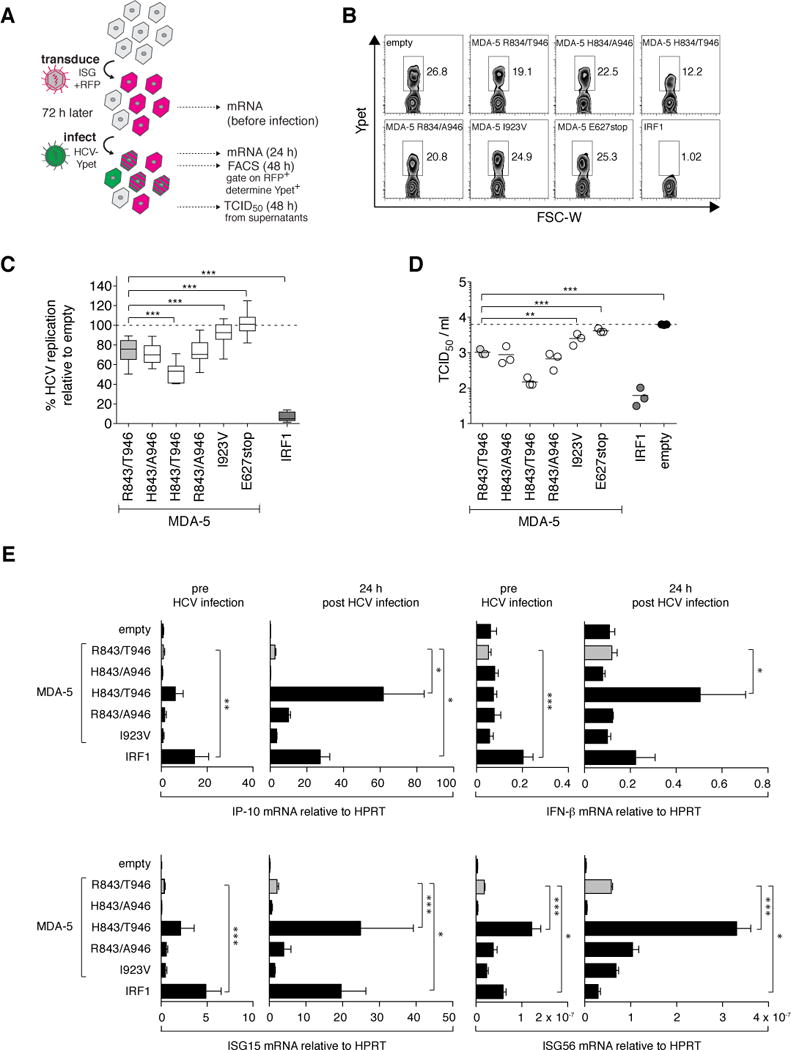
A) Workflow schematic: Huh-7.5 cells were transduced with lentiviral vectors expressing RFP and the indicated ISGs. MDA5 R843/T946, the most common allele combination (WT) in the European population, is highlighted in grey. Empty vector and two MDA-5 variants, I923V and E627stop, known to be functionally inactive, were used as negative controls. A construct coding for IRF1 served as positive control.72 hours after lentiviral transduction, cells were challenged with HCV Jc1 378-1-YPet. 48 h post infection, we harvested virus-containing supernatants for TCID50 determination, and cells for FACS analysis, or mRNA extraction and expression studies. B) FACS plots of one representative experiment. C. HCV replication efficiency is defined as the percentage of Ypet+ cells in the ISG-expressing (RFP+) population, normalized to the negative control (empty vector), which is set as 100%. Data of at least n = 6 independent experiments are depicted as means ± SD. D) TCID50 values from cell supernatants from n=3 experiments. E) Prior and 24 h post infection with HCV, cellular RNA was isolated and expression levels of the indicated genes were analysed by quantitative rt-PCR. Target gene expression was determined relative to the HPRT gene and is shown as means + SEM of n = 3. p-values were determined using the WT (R843/T946) as reference value. * p<0.05, ** p<0.01 and *** p<0.001.
Discussion
Our data present the first genetic evidence for a role of MDA-5 in the natural course of HCV infection. Our findings strongly argue in favor of an HCV-protective haplotype which is present in ~ 5% of the European population and which encodes an MDA-5 variant with enhanced functional activity.
Studies implicating the RLH pathway in HCV recognition (15, 16, 18, 26) so far suggested RIG-I as the primary RLH immune sensor for HCV. Interestingly, West Nile virus, an RNA virus also in the Flaviviridae family, is sensed by both RIG-I and MDA-5 (27). Furthermore, detailed studies examining specificity and potency of antiviral proteins showed that overexpressed MDA-5 is as potent as RIG-I in inhibiting HCV-replication (17). Knock-down of MDA-5 partially rescues HCV replication suppressed via IFN-α in the RIG-I negative Huh-7.5 cell line (28) and leads to reduced ISG-induction in RIG-I competent HepG2 cells (29). Overexpression of paramyxovirus V proteins that were shown to engage and inhibit MDA-5 but not RIG-I lead to increased HCV replication and spread in primary human fetal liver cells (30, 31). It is also important to note that the HCV NS3-4A serine protease has evolved to interfere with the RLH pathway at the level of MAVS, the adapter for both, RIG-I and MDA-5 (16). In the present study we see that overexpression of a functionally active MDA-5 is a prerequisite for induction of ISGs upon HCV infection in RIG-I deficient Huh-7.5 cells (figure 3E). Together, these findings implicate MDA-5 as an important determinant in the outcome of HCV infection.
The molecular pattern sensed by MDA-5 is still incompletely understood but studies indicate that the viral ligands for MDA-5 consist of long, higher-order dsRNA or dsRNA replication intermediates (11, 32–34). Cell culture studies have defined distinct molecular patterns in the genomic RNA of HCV that can be sensed by RIG-I and PKR (reviewed in (15)). While these studies showed, that in vitro transcribed HCV RNA itself does not require MDA-5 to induce type-I interferon, dsRNA replicative intermediates of HCV that accumulate at later times during the replication cycle carry the characteristics to be bona fide ligands for MDA-5. Further studies however are required to define the exact nature of the RNA species activating MDA-5 during HCV infection.
An early study analysed the SNPs rs1990760 and rs3747517 of MDA-5 separately and used a plasmid-construct coding for arginine 843/alanine 946 as the “wild type” source to generate their MDA-5 variants via point mutations. It thereby missed an effect of the haplotype coding for H843/T946 and disregarded a significant effect of theses SNPs on protein function (25). Later studies therefore assumed that these SNPs influence expression rather than function (22). However, a study comparing the expression levels for each allele separately in heterozygotes convincingly showed that rs1990760 does not influence MDA-5 promoter activity (35). Our results show that overexpression of the H843/T946 MDA-5 variant leads to a significantly higher induction of the interferon-stimulated gene products. In addition, compared to wild type MDA-5 the H843/T946 MDA-5 variant leads to significantly stronger inhibition of HCV replication in Huh-7.5 cells. This suggests that protein activity itself, and not elevated expression, is the reason for improved clearance of HCV infection in these haplotypes.
Novel insight into the structure of ligand-bound CARD-less MDA-5 and of its homologue RIG-I (36–40) might explain our findings. AA 843 is situated within one of the most striking structural features of the RLH family, the elbow-like pincer motif. The pincer structure is proposed to open and close upon RNA binding thereby altering the orientation of the CTD to the helicase domains. In addition, AA 843 is directly adjacent to residues forming the head surface of the putative contact interface between MDA-5 monomers within the recently described MDA-5 filaments that form along dsRNA (39, 41). AA 946 of MDA-5 is located at the center of a loop within the C-terminal domain that, in RIG-I, is directly in contact with its RNA ligand (supplementary figure 2). The depletion of the CTD-loop in MDA-5 leads to reduced signaling capacity, even though the affinity to dsRNA ligands is not reduced (39). The pincer motif and the CTD loop were described as two structures in RLHs that make large movements upon activation required to adjust the fitting of the receptors to their RNA-ligands. It is feasible that the two changes in H843/T946 MDA-5 at position 843 and 946 synergistically increase sensing of the viral RNA and/or improve transduction of the signal to downstream signaling molecules, resulting in a more efficient innate immune response that ultimately clears HCV infection in patients.
However, MDA-5 with increased activity might come at a cost. Indeed, several studies have identified T at rs1990760 in MDA-5 as a risk allele for type 1 diabetes noting LD with rs3747517 (22, 23). Furthermore, rare loss-of-function variants of MDA-5 are protective against type I diabetes (24) and there is increasing evidence that enterovirus infection, which is sensed by MDA-5, plays an important role in triggering type I diabetes (42). A recent paper by Rice et al. found gain-of-function mutations in MDA-5 causing Aicardi-Goutières syndrome and clinical related syndromes. In these cases so far unidentified endogenous MDA-5 ligands induce a spontaneous chronic induction of ISGs that are supposed to be causative for the clinical phenotype of a severe inflammatory multi-organ disease (43). We saw a slight ISG stimulation upon overexpression of MDA5 H843/T946 even in the absence of infection or poly I:C in HEK293 and Huh-7.5 cells. Interestingly, Rice et al. argue that baseline ISG-induction due to overexpression of MDA-5 variants in the absence of added ligands is in fact not completely ligand-independent but rather depends on endogenous RNA ligands. Their argument being that the additional introduction of mutations disrupting the putative RNA binding site blocks this baseline activation (43).
It is therefore possible that H843/T946 is an overactive MDA-5 variant that is beneficial for HCV clearance, but might be a risk factor for autoimmune diseases such as diabetes type I. We speculate that a strong response to cytoplasmic viral RNA, or baseline activation by endogenous ligands though an advantage in the fight against the persistence of HCV, might trigger autoimmunity by increased bystander activation or induction of apoptosis. Further studies will be needed to see if the strong effect of the H843/T946 MDA-5 variant on HCV clearance can be generalized to other viral infections sensed by MDA-5, and is conversely associated with increased risk to autoimmune diseases beyond type I diabetes as suggested by recent studies (44).
Supplementary Material
Acknowledgments
We would like to thank Dr. Müller, Dr. Viazov, Dr. Roggendorf and the other members and organizers of the German Kompetenznetz Hepatitis for access to their sample collection and databases and all involved patients and their physicians for enabling this study. We are grateful to Paul Bieniasz, The Rockefeller University, for providing us with the SCRPSY lentivirus vector. We thank Dr. E. Albert for carefully reading the manuscript.
Grant support
The study was supported by DFG RO 2525/3-1 and RO 2525/5-1 to S.R., DFG GK 1202 to A.S., S.E. and S.R., by LMUexcellent (CIPSM 114, research professorship) and BayImmuNet to S.E., by National Institutes of Health grant AI091707 to C.M.R., and by The Rockefeller University Women and Science grant to M.D.C. T. Berg is supported by the German Competence Network for Viral Hepatitis (Hep-Net) through BMBF Grant No. 01 KI 0437, by the EU-Vigilance network of excellence combating viral resistance (VIRGIL, Project No. LSHM-CT-2004-503359) and by BMBF Grant No. 01KI0787.
List of Abbreviations
- AA
amino acid
- C
controls
- Cardif
CARD adaptor inducing IFN-beta
- CH
chronic hepatitis
- DHX58
DEXH (Asp-Glu-X-His) box polypeptide 58
- DDX58
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58
- FRET
fluorescence resonance energy transfer
- HCV
hepatitis C virus
- IFIH1
interferon-induced with helicase C domain 1
- IFN
interferon
- IPS-1
IFN-beta promoter stimulator-1
- IRF
interferon regulatory factor
- LD
linkage disequilibrium
- Lgp2
laboratory of genetics and physiology 2
- MAF
minor allele frequency
- MAVS
mitochondrial antiviral signaling protein
- MDA-5
melanoma differentiation-associated gene 5
- NFkB
nuclear factor of kappa light polypeptide gene enhancer in B-cells
- NS3/4A
nonstructural protein 3/4A
- OR
odds ratio
- PRR
pattern-recognition receptor
- RIG-I
retinoic acid inducible gene-I
- RLH
RIG-I like helicase, SNP, single nucleotide polymorphism
- SRH
spontaneously resolved hepatitis
- VISA
virus-induced signaling adapter
Contributor Information
Franziska Hoffmann, Email: franziska.hoffmann@med.uni-muenchen.de.
Andreas Schmidt, Email: aschmidt@mpiib-berlin.mpg.de.
Meike Dittmann Chevillotte, Email: mchevillot@mail.rockefeller.edu.
Christian Wisskirchen, Email: chr.wisskirchen@gmail.com.
Johannes C. Hellmuth, Email: johannes.hellmuth@med.uni-muenchen.de.
Simone Willms, Email: simone.willms@googlemail.com.
Rachel H. Gilmore, Email: Rachel.h.gilmore@gmail.com.
Jürgen Glas, Email: jglas@ukaachen.de.
Matthias Folwaczny, Email: mfolwa@dent.med.uni-muenchen.de.
Tobias Müller, Email: tobias.mueller@medizin.uni-leipzig.de.
Thomas Berg, Email: thomas.berg@medizin.uni-leipzig.de.
Ulrich Spengler, Email: ulrich.spengler@ukb.uni-bonn.de.
Karen Fitzmaurice, Email: kfitzmau@tcd.ie.
Dermot Kelleher, Email: kellehdp@tcd.ie.
Nicole Reisch, Email: Nicole.Reisch@med.uni-muenchen.de.
Charles M. Rice, Email: ricec@mail.rockefeller.edu.
Stefan Endres, Email: endres@lmu.de.
Simon Rothenfusser, Email: simon.rothenfusser@med.uni-muenchen.de.
References
- 1.Romero-Gomez M, Eslam M, Ruiz A, Maraver M. Genes and hepatitis C: susceptibility, fibrosis progression and response to treatment. Liver Int. 2011;31:443–460. doi: 10.1111/j.1478-3231.2011.02449.x. [DOI] [PubMed] [Google Scholar]
- 2.Selvarajah S, Tobler LH, Simmons G, Busch MP. Host genetic basis for hepatitis C virus clearance: a role for blood collection centers. Curr Opin Hematol. 2010;17:550–557. doi: 10.1097/MOH.0b013e32833e7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 4.Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 5.Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 6.Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, Kidd J, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Metz P, Reuter A, Bender S, Bartenschlager R. Interferon-stimulated genes and their role in controlling hepatitis C virus. J Hepatol. 2013;59:1331–1341. doi: 10.1016/j.jhep.2013.07.033. [DOI] [PubMed] [Google Scholar]
- 8.Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2012;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 10.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 11.Pichlmair A, Schulz O, Tan CP, Rehwinkel J, Kato H, Takeuchi O, Akira S, et al. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J Virol. 2009;83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, Wenzel M, Hoffmann FS, et al. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci U S A. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loo YM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt A, Endres S, Rothenfusser S. Pattern recognition of viral nucleic acids by RIG-I-like helicases. J Mol Med. 2011;89:5–12. doi: 10.1007/s00109-010-0672-8. [DOI] [PubMed] [Google Scholar]
- 15.Horner SM, Gale M., Jr Regulation of hepatic innate immunity by hepatitis C virus. Nat Med. 2013;19:879–888. doi: 10.1038/nm.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 17.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sumpter R, Jr, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, et al. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenny-Walsh E. Clinical outcomes after hepatitis C infection from contaminated anti-D immune globulin. Irish Hepatology Research Group. N Engl J Med. 1999;340:1228–1233. doi: 10.1056/NEJM199904223401602. [DOI] [PubMed] [Google Scholar]
- 20.Horwitz JA, Dorner M, Friling T, Donovan BM, Vogt A, Loureiro J, Oh T, et al. Expression of heterologous proteins flanked by NS3-4A cleavage sites within the hepatitis C virus polyprotein. Virology. 2013;439:23–33. doi: 10.1016/j.virol.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 22.Liu S, Wang H, Jin Y, Podolsky R, Reddy MV, Pedersen J, Bode B, et al. IFIH1 polymorphisms are significantly associated with type 1 diabetes and IFIH1 gene expression in peripheral blood mononuclear cells. Hum Mol Genet. 2009;18:358–365. doi: 10.1093/hmg/ddn342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smyth DJ, Cooper JD, Bailey R, Field S, Burren O, Smink LJ, Guja C, et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet. 2006;38:617–619. doi: 10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- 24.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shigemoto T, Kageyama M, Hirai R, Zheng J, Yoneyama M, Fujita T. Identification of loss of function mutations in human genes encoding RIG-I and MDA5: implications for resistance to type I diabetes. J Biol Chem. 2009;284:13348–13354. doi: 10.1074/jbc.M809449200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fredericksen BL, Keller BC, Fornek J, Katze MG, Gale M., Jr Establishment and maintenance of the innate antiviral response to West Nile Virus involves both RIG-I and MDA5 signaling through IPS-1. J Virol. 2008;82:609–616. doi: 10.1128/JVI.01305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci U S A. 2009;106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Israelow B, Narbus CM, Sourisseau M, Evans MJ. HepG2 cells mount an effective antiviral interferon-lambda based innate immune response to hepatitis C virus infection. Hepatology. 2014 doi: 10.1002/hep.27227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andrus L, Marukian S, Jones CT, Catanese MT, Sheahan TP, Schoggins JW, Barry WT, et al. Expression of paramyxovirus V proteins promotes replication and spread of hepatitis C virus in cultures of primary human fetal liver cells. Hepatology. 2011;54:1901–1912. doi: 10.1002/hep.24557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Motz C, Schuhmann KM, Kirchhofer A, Moldt M, Witte G, Conzelmann KK, Hopfner KP. Paramyxovirus V proteins disrupt the fold of the RNA sensor MDA5 to inhibit antiviral signaling. Science. 2013;339:690–693. doi: 10.1126/science.1230949. [DOI] [PubMed] [Google Scholar]
- 32.Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Triantafilou K, Vakakis E, Kar S, Richer E, Evans GL, Triantafilou M. Visualisation of direct interaction of MDA5 and the dsRNA replicative intermediate form of positive strand RNA viruses. J Cell Sci. 2012;125:4761–4769. doi: 10.1242/jcs.103887. [DOI] [PubMed] [Google Scholar]
- 35.Zouk H, Marchand L, Polychronakos C. Study of transcriptional effects in Cis at the IFIH1 locus. PLoS One. 2010;5:e11564. doi: 10.1371/journal.pone.0011564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 37.Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M, Jr, Patel SS, Marcotrigiano J. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu B, Peisley A, Richards C, Yao H, Zeng X, Lin C, Chu F, et al. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 40.Peisley A, Lin C, Wu B, Orme-Johnson M, Liu M, Walz T, Hur S. Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proc Natl Acad Sci U S A. 2011;108:21010–21015. doi: 10.1073/pnas.1113651108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berke IC, Yu X, Modis Y, Egelman EH. MDA5 assembles into a polar helical filament on dsRNA. Proc Natl Acad Sci U S A. 2012;109:18437–18441. doi: 10.1073/pnas.1212186109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tauriainen S, Oikarinen S, Oikarinen M, Hyoty H. Enteroviruses in the pathogenesis of type 1 diabetes. Semin Immunopathol. 2011;33:45–55. doi: 10.1007/s00281-010-0207-y. [DOI] [PubMed] [Google Scholar]
- 43.Rice GI, Del Toro Duany Y, Jenkinson EM, Forte GM, Anderson BH, Ariaudo G, Bader-Meunier B, et al. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat Genet. 2014;46:503–509. doi: 10.1038/ng.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cen H, Wang W, Leng RX, Wang TY, Pan HF, Fan YG, Wang B, et al. Association of IFIH1 rs1990760 polymorphism with susceptibility to autoimmune diseases: a meta-analysis. Autoimmunity. 2013;46:455–462. doi: 10.3109/08916934.2013.796937. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


