Figure 5.
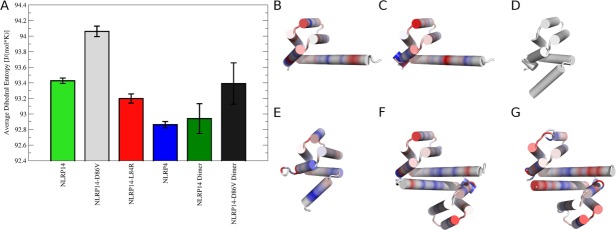
Conformational entropies of NLRPs: (A) Dihedral entropies were calculated over backbone torsions and averaged over the whole protein. Monomeric NLRPs in open form (NLRP14, NLRP14-D86V) show highest entropy, thus showing the broadest conformational ensemble. Conformational space is restricted for closed state monomers (NLRP14-L84R, NLRP4) and dimeric NLRPs. (B–G) Differences in conformational entropies in NLRPs: Residue-wise dihedral entropies were mapped to the starting structures in cartoon representation relative to the stabilized mutant NLRP14-L84R. Differences are shown on a color gradient from red (more flexible, +15 J (mol K−1)−1) over white (no change) to blue (more rigid, −15 J (mol K−1)−1). B) Monomeric native NLRP14 and (C) monomeric mutant NLRP14-D86V show mobilized kink regions in the elongated α5/6 stem helix. (D) Reference structure NLRP14-L84R (completely white). (E) NLRP4 is overall more rigid compared to NLRP14-L84R. Dimeric systems of native NLRP14 (F) and mutant NLRP14-D86V (G) show local rigidification in the dimerization interface. Dimeric NLRP14-D86V has still preserved residual flexibility in the α6 helix.
