Figure 2.
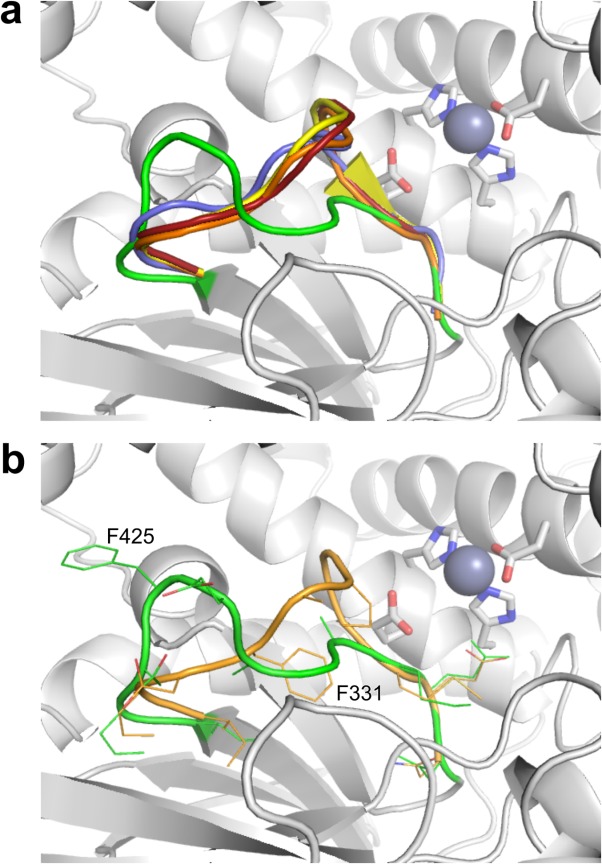
Comparison of the GAMEN motif loop conformation in several representative M1 aminopeptidase structures. The structures of ERAP1 open (PDB id: 3MDJ, blue), ERAP1 closed (PDB id: 2YD0, yellow), ERAP2 (PDB id: 4JBS, orange), and APN (PDB id: 4FYS, red) were superimposed on the IRAP crystal structure (green) using the catalytic site residues. (a) The different conformations of the GAMEN motif loops for the proteins colored as above. The rest of the IRAP structure is shown as a gray cartoon with the active site residues shown as rods colored by atom type. (b) Detailed representation of the IRAP and ERAP2 GAMEN motif loops. The two proteins are displayed as in (a) but with the residues of the loop shown as sticks colored by atom type. Displacement of the loop between the equivalent residues Ile422/328 and Met430/336 is substantial, with the difference in the side-chains of Phe425/331 exceeding 10 Å. An interactive view is available in the electronic version of the article.
