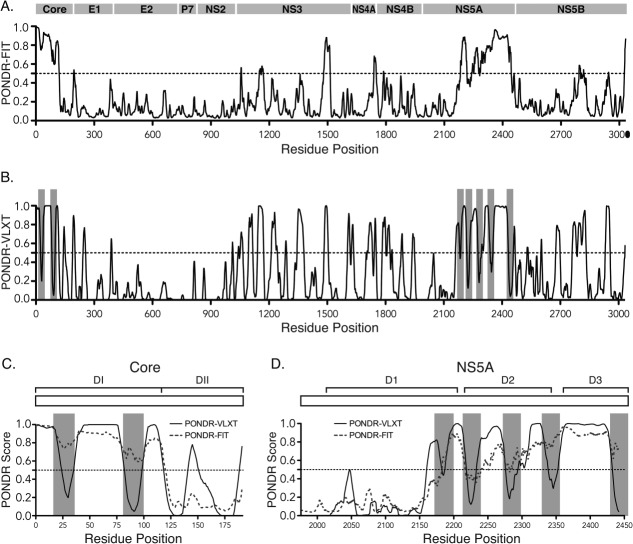
Figure 1.
Sequence-based prediction of intrinsic disordered regions (IDRs) in the HCV polyprotein. (A) The PONDR®-FIT meta-predictor was used to estimate the level of disorder in the HCV JFH1 polyprotein. Higher PONDR scores indicate a higher propensity to be unstructured. Bar above the graph represents the HCV polyprotein. (B) PONDR®-VLXT scores were used along with the α-MoRF-pred tool to identify α-helical Molecular Recognition Features (α-MoRFs) in the HCV JFH1 polyprotein sequence. Grey blocks indicate the positions of predicted α-MoRFs. (C and D) PONDR®-FIT and –VLXT scores for HCV Core (C) and NS5A (D). Schematics of the Core and NS5A proteins are shown above the graphs. Brackets indicate domains.132