Figure 5.
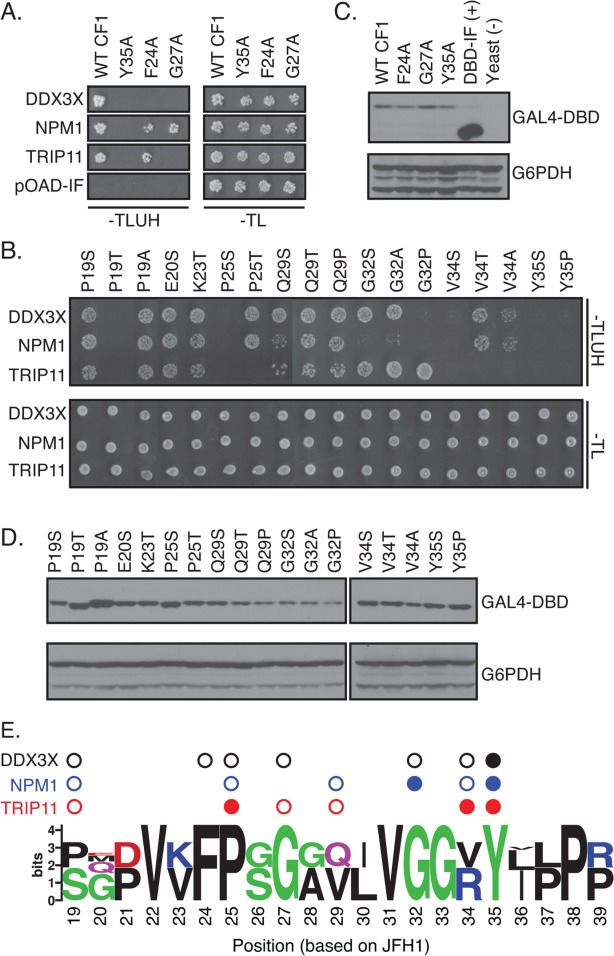
Distinct amino acids in MoRF1 are required for binding to cellular proteins. (A and B) Single residue substitutions in Core MoRF1 disrupt the binding to cellular proteins. (A) Substitutions previously shown to disrupt binding of Core to DDX3X71 were assayed for their impact on binding to cellular proteins using the Y2H assay. Top panel shows growth on Y2H selection medium whereas bottom panel shows growth on control medium. pOAD-IF is a control plasmid, in which URA3 is in frame with the GAL4 activation domain and no human gene is present. (B) Single residue substitutions were introduced into Core MoRF1 by site-directed mutagenesis and tested for their effect on binding to cellular proteins using the Y2H assay. Top panel, Y2H selection medium; bottom panel; growth control medium. (C and D) Western blots to confirm expression of mutant Core fragments. Upper blot was probed with anti-GAL4 DNA binding domain antibody (GAL4-DBD); lower blot was probed with anti-glucose-6-phosphate dehydrogenase antibody (G6PDH) as a loading control. (E) Summary of the effect of HCV Core MoRF1 substitutions on binding to human proteins. Web Logo of Core MoRF1 was derived from alignment of the six HCV genotypes and eight representative NPHV isolates. Circles indicate substitutions that disrupted binding to DDX3X (black), NPM1 (blue), or TRIP11 (red). Shaded circles indicate multiple substitutions at that position disrupted binding. All experiments were performed at least twice in independent replicates; representative results are shown.
