Fig. 1.
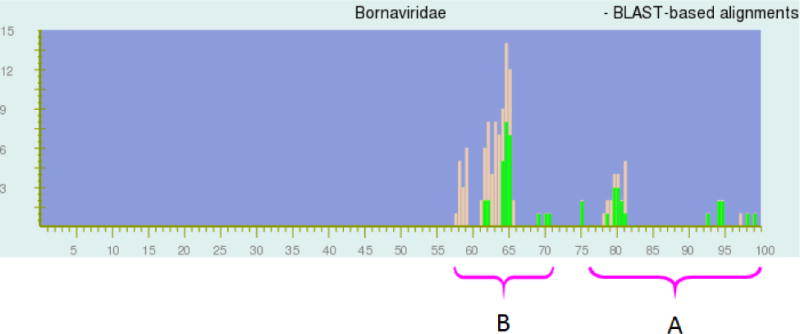
Distribution of pairwise identities among complete sequences of 16 viruses in the family Bornaviridae. The NCBI PASC alignment tool automatically assigns peak colors (brown, green) based on NCBI virus taxonomy. Regions represent genome pairs belonging to the same species (A) and to different species but to the same genus (B), respectively. Since most of the sequences used for PASC analysis are currently not classified in NCBI taxonomy the same way as proposed in this paper, the colors in this screenshot do not follow the grouping, thereby visually demonstrating that the current bornavirus taxonomy is ill-advised. For instance, bornavirus genomes EU781967 and FJ620690 are 97% identical and therefore should belong to the same species in one genus. However, in the current NCBI taxonomy, EU781967 is assigned to the species Borna disease virus in the genus Bornavirus, whereas FJ620690 is assigned to an unclassified species in an unclassified genus in the family Bornaviridae. Therefore, since the NCBI taxonomy tool does not assign both sequences to the same species or the same genus, the bar at 97% in the current PASC figure is colored brown rather than green. X-axis, percentage of identity; y-axis, number of genome pairs
