Fig. 3.
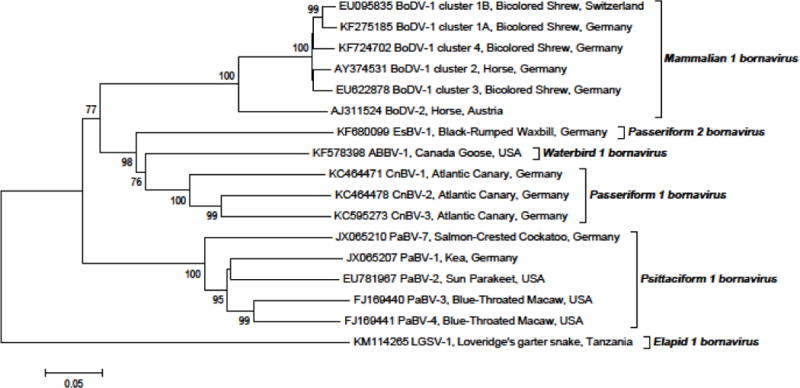
Phylogenetic tree of a 1824-nt stretch of 17 selected nucleic acid sequences coding for N, X, and P proteins and the N/X intergenic region of bornaviruses. Phylogenetic neighbor-joining analysis was conducted using the MEGA5 program [55]. The evolutionary distances were computed using the Kimura 2-parameter model. Bootstrap resampling analysis with 1,000 replicates was employed; percentages ≥60% are shown next to the branches.
