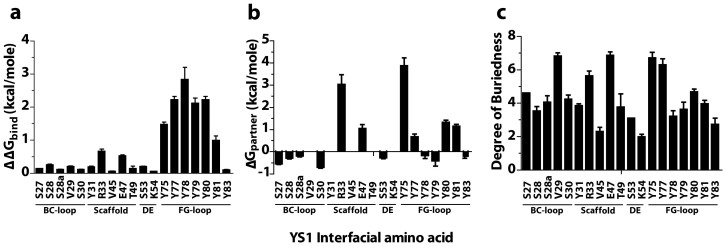
Figure 1. Computational simulations predict the critical residues of YS1 at the YS1-MBP binding interface.
(a) DrugscorePPI webserver prediction of changes in binding free energy (ΔΔGbind) upon alanine mutation. (b) Robetta prediction of the effect of alanine mutation on mutated protein complex partner in isolation (ΔGpartner). Residues V29, Y31, V45, T49 and K54 were omitted as Robetta predictions failed to identify them as being present in 50% of the predicted binding complexes. (c) DrugscorePPI webserver prediction of the degree of buriedness of the interfacial residues at the YS1-MBP interface. Results are the mean ± s.e.m. of 6-8 pairs of randomly selected monobody-MBP complexes.