Table 1. Comparison of experimentally determined kinetic constants for mutations at critical scaffold residues as determined by scanning alanine mutagenesis simulations.
| DrugscorePPI | Robetta | Single-molecule force spectroscopy | Suface plamon resonance | ||||
|---|---|---|---|---|---|---|---|
| sample | ΔΔGbind (kcal/mol) | ΔΔGpartner (kcal/mol) |
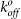 (× 10−3s−1) (× 10−3s−1) |
xβ(nm) | kon(× 105M−1s−1) |
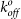 (× 10−3s−1) (× 10−3s−1) |
Kd (nM) |
| Wild-type | 0.24 ± .06 | 0.42 ± .03 | 4.4 ± 0.7 | 40.6 ± 3.2 | 100 ± 18 | ||
| E47A | .53 ± .03 | 1.01 ± .18 | 0.76 ± .39 | 0.46 ± .05 | 3.6 ± 0.3 | 38.7 ± 15 | 101 ± 41 |
| R33A | .67 ± .06 | 3.04 ± .44 | N.B. | — | N.B. | — | — |
N.B. represents no binding. This No binding is defined as having a binding frequency under 5% for single molecule force spectroscopy or displaying an <5 RU response up to 1 μM concentration in SPR. Data represent the mean ± s.e.m. of at least three independent experiments.
