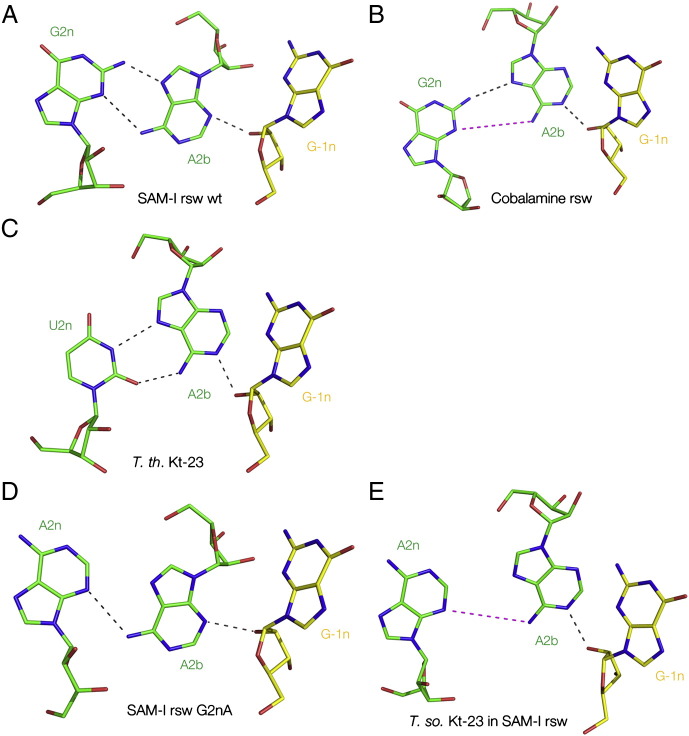
Fig. 7.
Hydrogen bonding between the 2b·2n pair and the O2′ of − 1n in N3 and N1-class K-turn structures. Hydrogen bonds are indicated by gray broken lines. Distances too long to be considered hydrogen bonded are colored magenta.
A. The SAM-I riboswitch K-turn [4], a standard N3-class K-turn.
B. The cobalamine riboswitch K-turn [7], an N1-class K-turn. Note that the reorientation of the A2b nucleobase results in a A2b N6–G2n N3 N–N distance that is too long for stable hydrogen bond formation.
C. The T. thermophilus Kt-23 K-turn [36]. This K-turn has a uridine at the 2n position, making a standard Hoogsteen basepair with A2b. The latter accepts a proton from G − 1n O2′ at the N1 position, making this an N1-class K-turn.
D. The SAM-I riboswitch k-turn with a G2nA substitution, making an A·A pair at the 2b·2n position. This forms a stable K-turn structure in the context of the riboswitch, for which the crystal structure has been determined [38]. A2b accepts a proton from G − 1n O2′ at the N3 position, and there is a hydrogen bond between A2b N6 and A2n N3. This is a standard N3 structure.
E. The T. solenopsae Kt-23 K-turn, engineered into the SAM-I riboswitch. This rare K-turn sequence naturally has an adenine at the 2n position. But in the context of the riboswitch, this K-turn adopts the N1 structure [26]. A2b accepts a proton from G − 1n O2′ at the N1 position, and there is no hydrogen bond between A2b N6 and A2n N3.