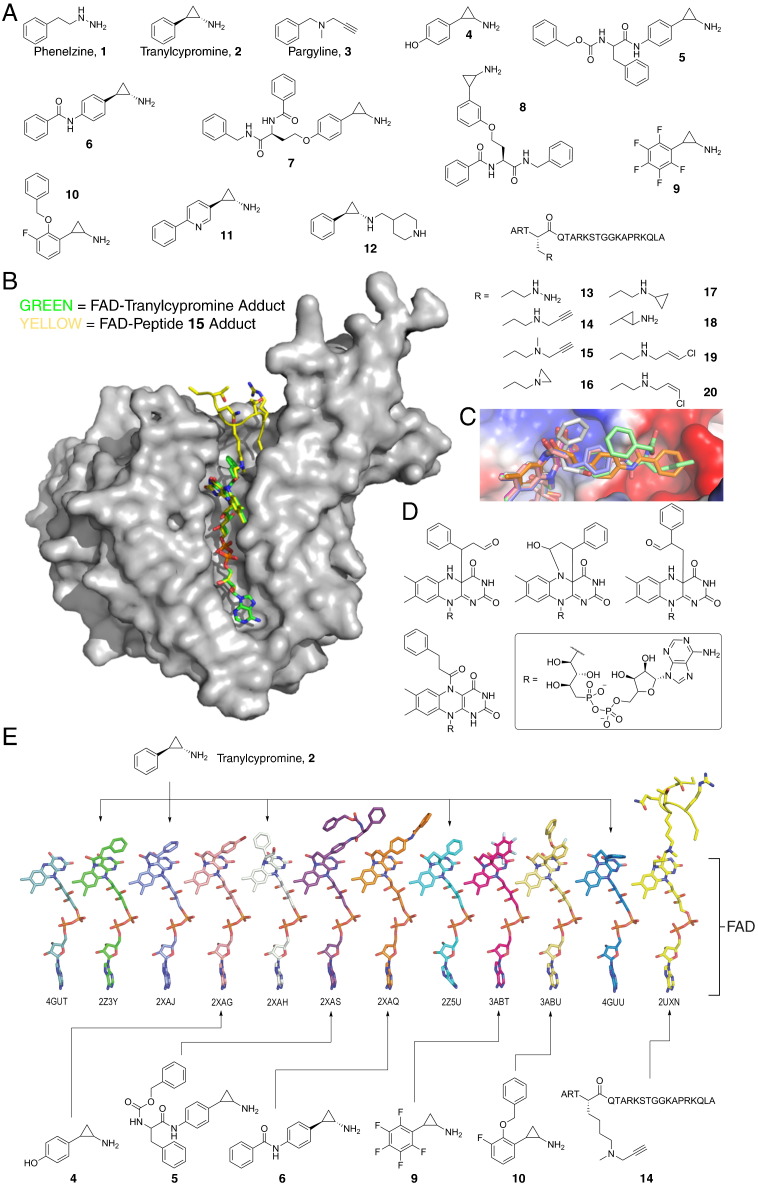
Fig. 2.
Mechanism-based inhibitors of the KDM1 subfamily. (A) Structures of representative mechanism-based KDM1 inhibitors. The MAO inhibitors phenelzine 1, tranylcypromine 2 and pargyline 3 were among the first reported inhibitors of KDM1, which led to the development of KDM1-selective analogues. (B) Views from X-ray crystal structures of tranylcypromine (2) and peptidic inhibitor (14) cross-linked to FAD in the active site of KDM1A (PDB IDs: 2Z3Y[63] and 2UXN[155] respectively). Views from both structures are overlaid. (C) Views from crystal structures of tranylcypromine analogues (FAD adducts) in the active site of KDM1A (PDB IDs: 2XAJ, 2XAG, 2XAH, 2XAS and 2XAQ2XAS2XAQ) [61]. The analogues protrude into the substrate binding pocket. (D) Proposed structures of adducts formed by reaction of tranylcypromine with FAD in the active site of KDM1s. There is evidence for three of the structures from crystallographic analyses (see section E). (E) Structures of FAD adducts of mechanism-based inhibitors bound in the active sites of KDM1s (views of structures are from PDB IDs: 2Z3Y[63], 2XAJ[61], 2XAG[61], 2XAH[61], 2XAS[61], 2XAQ[61], 2Z5U[63], 3ABT[66], 3ABU, 4GUU[156] and 2UXN[155]).