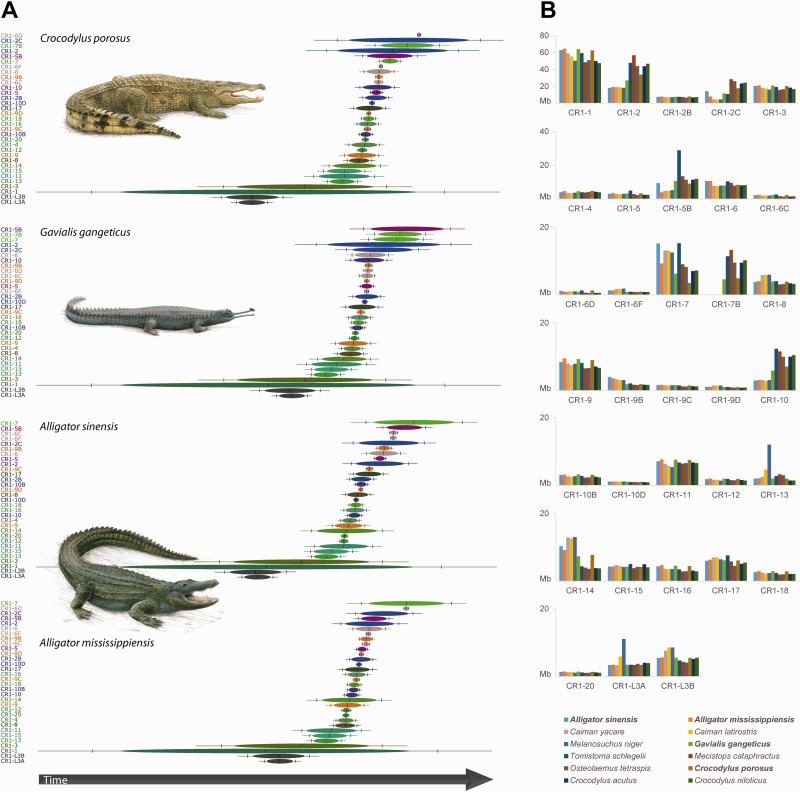
Fig. 2.—
Activity of CR1 elements in 12 crocodilian genomes. (A) Estimates of CR1 activity in genome assemblies of saltwater crocodile, gharial, Chinese alligator, and American alligator using the TinT model (Kriegs et al. 2007; Churakov et al. 2010). Congruent successions of predicted activities of merged CR1 subfamilies (see supplementary figure S1, Supplementary Material online, for unmerged TinT patterns) between the four genomes suggest that the core activity periods of most subfamilies lie in the common ancestor of crocodilians. Normal distributions of activity period estimations are shown as ovals (Churakov et al. 2010). (B) Comparison of CR1-derived base pair quantities in four assembled genomes (bold names) and eight unassembled survey sequence libraries. All CR1 subfamilies with the exception of CR1-7B appear to be present across the breadth of the sampled crocodilian species diversity. Nevertheless, quantitative differences within several subfamilies suggest differential activities in crocodilian genomes; for example, an extended activity of CR1-2 in Longirostres (all crocodilians except alligators and caimans (Harshman et al. 2003)) and of CR1-5B in T. schlegelii. Note that some of the lineage-specific differences in CR1 quantities might either be accounted to potential differences in rate of neutral sequence evolution or cryptic CR1 subfamilies that were not identified in our CR1 predictions (as they are based on the genome assemblies of saltwater crocodile, gharial, and American alligator). English names of the 12 genomes are (in the order of their appearance in the legend) Chinese alligator, American alligator, Yacare caiman, broad-snouted caiman, black caiman, gharial, false gharial, slender-snouted crocodile, dwarf crocodile, saltwater crocodile, American crocodile, and Nile crocodile.