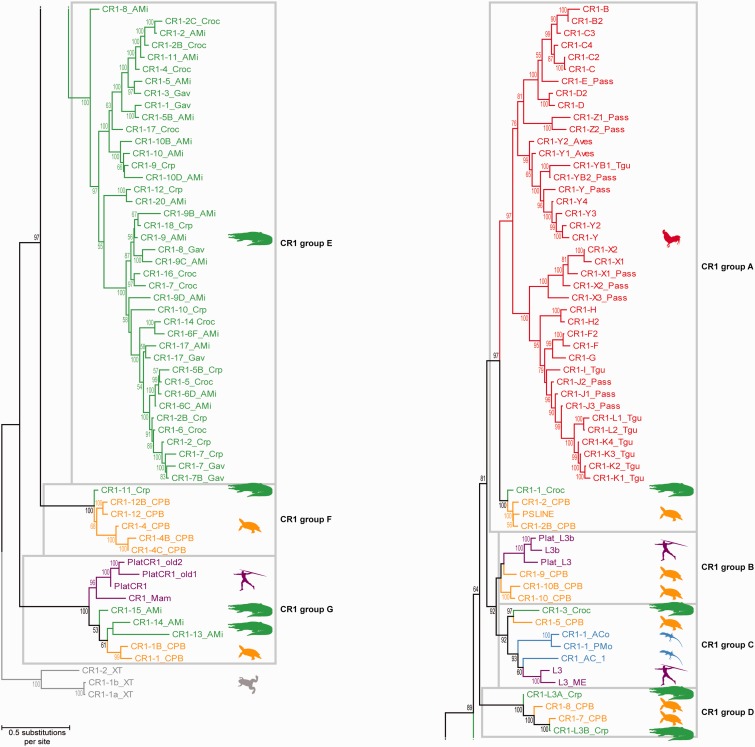
Fig. 3.—
Phylogenetic relationships among amniote CR1 elements. Maximum likelihood nucleotide sequence analysis (RAxML 8.0.0 (Stamatakis et al. 2008), GTRCAT model, 1,000 bootstrap inferences) of part of the ORF2 RT domain + 3′-UTR of all CR1 subfamily consensus sequences available in RepBase (Jurka et al. 2005) suggests a complex tree topology of avian (red), crocodilian (green), turtle (orange), lepidosaurian (blue) and mammalian (purple) CR1 elements. Given this topology and the parsimonious assumption that CR1 retroposons are inherited vertically, we infer a minimum number of seven ancient CR1 lineages (groups A–G; see also supplementary table S3, Supplementary Material online, for a group classification of all analyzed CR1 subfamilies) that were present in the common ancestor of amniotes. The tree was rooted using amphibian CR1 subfamilies from Xenopus as outgroup, as they were the only nonamniote CR1 elements alignable to ingroup CR1s on the nucleotide level. Note that, in contrast to the merged CR1 subfamily definitions of figures 1 and 2, we included all CR1 consensus sequences generated in the individual TE annotations of the saltwater crocodile, gharial, and American alligator genomes. Given the short internodes and high sequence similarity among the unmerged crocodilian CR1 subfamilies of group E, we refrain from renaming these CR1 subfamilies according to the topology of the present tree, until additional crocodilian genome assemblies are available. The present nomenclature of crocodilian CR1 subfamilies is therefore solely based on the UCLUST grouping of their consensus sequences (see Materials and Methods). Unlabeled nodes received a bootstrap support of less than 50%.