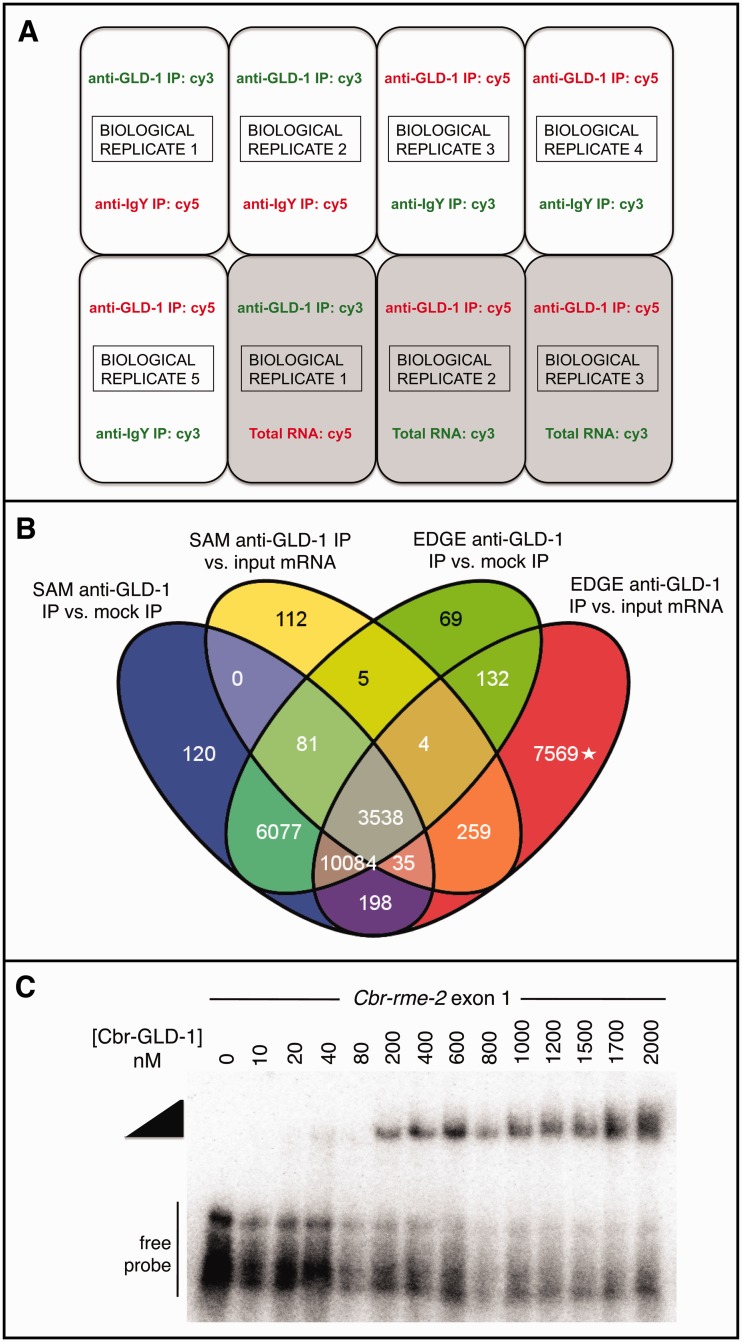
Fig. 2.—
Mircoarray experimental design and analysis to identify Cbr-GLD-1-associated RNAs. (A) Schematic of microarray design for two different expression comparisons, anti-GLD-1 IP mRNA versus mock anti-IgY IP mRNA and anti-GLD-1 IP mRNA versus total input mRNA. Dye swaps and biological replicates are incorporated. (B) Overlap of positive probes from the two microarray comparisons, anti-GLD-1 IP mRNA versus mock anti-IgY IP mRNA and anti-GLD-1 IP mRNA versus total input mRNA, each analyzed with two differential gene expression programs, SAM and EDGE. Values in each oval are the number of probes enriched in anti-GLD-1 IPs with FDRs of at most less than 2%. (The group of probes marked with the asterisk is enriched in both anti-GLD-1 IPs and total input mRNA.) In total, 3,538 probes were found enriched in common to all four data sets, representing 965 different Caenorhabditis briggsae protein coding genes. (C) The STAR domain of Cbr-GLD-1 can shift an RNA fragment of exon 1 RIP-chip target Cbr-rme-2, which contains multiple sequence motifs consistent with GLD-1 binding, in a concentration-dependent manner. Wedge, Cbr-GLD-1-dependent protein–RNA complex. A negative control RNA fragment from the 5′-UTR of Cbr-tra-2 does not shift at these concentrations (data not shown).