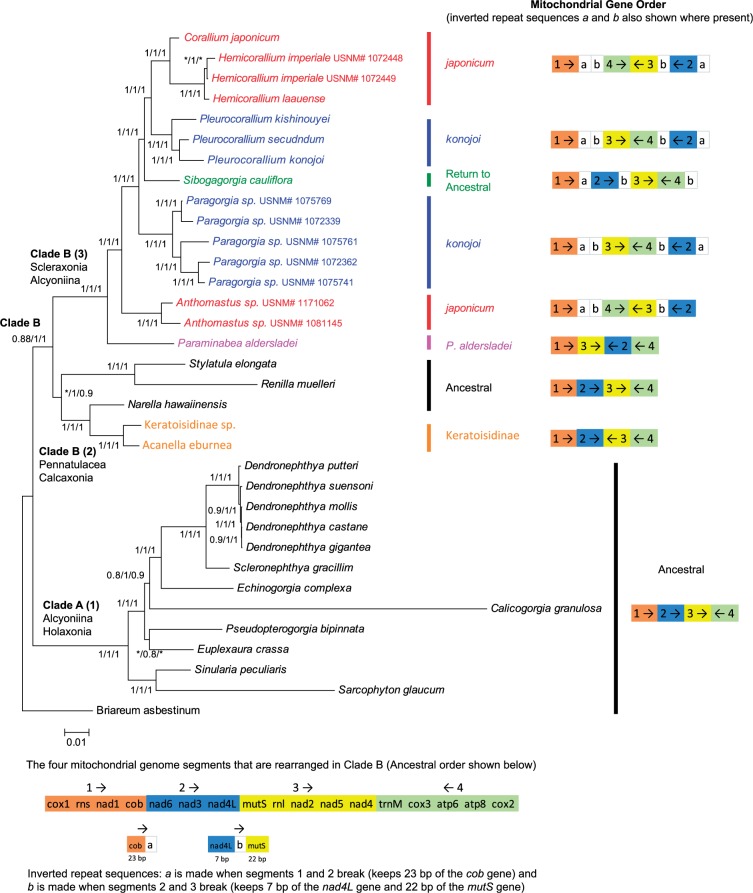
Fig. 5.—
Octocoral phylogenetic tree inferred by ML, based on all mitochondrial protein-coding genes and RNAs. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Tree topology inferred by Bayesian methods is identical except for Hemicorallium imperiale USNM# 1072449 branches with Hemicorallium imperiale USNM# 1072448 in the Bayesian topology and with Hemicorallium laauense in the ML topology. Branch values correspond to bootstrap support for ML (first) and Bayesian posterior probabilities (second) for the nonpartitioned data. The third branch value corresponds to bootstrap support for ML as determined by RAxML with the partitioned data. *Support values less than 0.70. Clade numbers are labeled to correspond to the clade designations in McFadden et al. (2006). Coloring corresponds to the different mitochondrial gene orders as shown to the right of the phylogeny. The corresponding genes in each numbered box are given in the bottom panel of the diagram.