Figure 4.
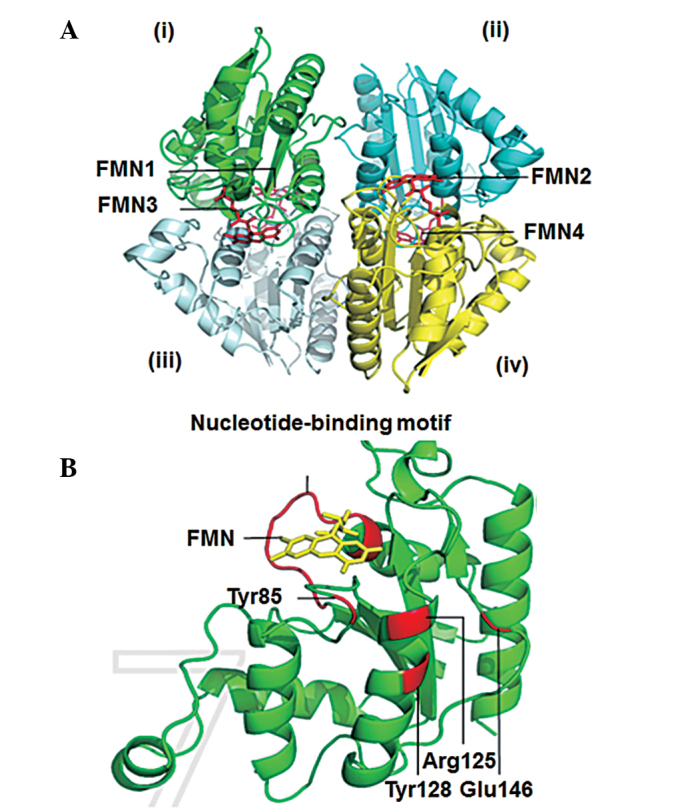
Three-dimensional structure model of ChrT predicted using the SWISS-MODEL program, according to the crystal structure of the ChrR enzyme from E. coli. (A) Crystal structure of the Serratia sp. CQMUS2 ChrT tetramer, which is formed by two dimers, with each monomer containing one binding site for FMN. One dimer is formed by monomers (i) and (ii), while the other dimer is formed by monomers (iii) and (iv). (B) Crystal structure of the nucleotide-binding motif showing FMN, as well as the Glu146, Tyr8, Arg125 and Tyr128 catalytic residues in ChrT. E. coli, Escherichia coli; FMN, flavin mononucleotide.
