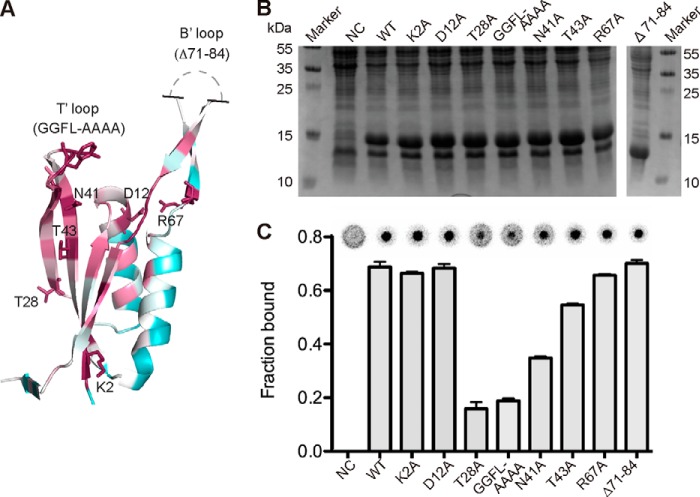
FIGURE 7.
Location of highly conserved amino acids in PstASA and their role in c-di-AMP binding. A, schematic representation of the PstASA monomer from the complex structure in the ConSurf representation (purple, high conservation; green, medium conservation; white, low conservation) with the mutated amino acids shown in stick representation. Unstructured region of the B′-loops is schematically indicated. B, Coomassie-stained gel of whole cell extracts prepared from E. coli BL21(DE3) strains containing the empty vector pET28b as negative control (NC), strains overproducing His-PstASA (WT), or different protein variants containing the amino acid substitutions as indicated above each lane. A protein marker was run alongside, and the molecular mass of the marker proteins is indicated in kDa on the left and right sides of the panel. C, c-di-AMP binding assays. DRaCALAs were performed with 32P-labeled c-di-AMP and the E. coli whole cell lysates described in B. Representative DRaCALA spots are shown, and the fraction bound was calculated as described previously, and the average values and standard deviation of two independent experiments are plotted.