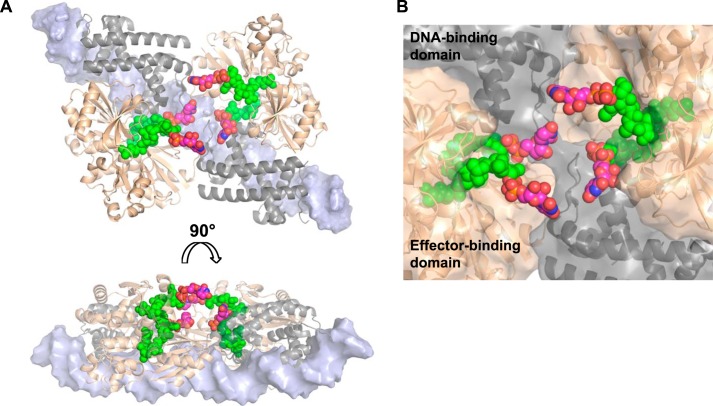
FIGURE 9.
Modeling of the AmpR·DNA complex bound to UDP-MurNAc-pentapeptide. A, based on the fitting of AphB (N100E) and dsDNA into our SAXS envelopes of the unliganded and UDP-MurNAc-pentapeptide-bound AmpR·DNA complexes, we built a molecular model of AmpR bound to the dsDNA molecules by superposing our crystal structure of the AmpR EBD homodimer bound to UDP-MurNAc-pentapeptide (shown in gold) onto the equivalent EBD dimers of the modeled AphB·DNA complex (shown in gray). A short, continuous fragment of double-stranded DNA (light blue) is shown docked onto the DNA-binding domains of AphB. UDP moieties (carbon atoms are shown in magenta, nitrogen in blue, oxygen in red, and phosphorus in yellow) were manually modeled on MurNAc-pentapeptide portions of the repressor that were visible in the electron density maps (green). B, a close-up of the central cavity predicted to exist within the AmpR tetramer showing how the UDP-MurNAc moieties of the bound repressor ligands may become positioned to interact with each other or with residues of the DNA-binding domains and/or EBDs to trigger conformational rearrangements in the quaternary structure of AmpR that cause it to bind the operator DNA in a manner that represses gene transcription.