FIGURE 2.
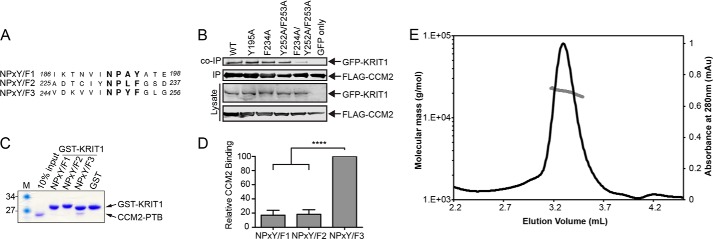
Mutations in KRIT1NPX(Y/F)2 and KRIT1NPX(Y/F)3 reduce the KRIT1 interaction with CCM2 and CCM2 preferentially interacts with KRIT1NPX(Y/F)3in vitro. A, sequence alignment of the three KRIT1 NPX(Y/F) motifs. Residue numbers are indicated. B, co-immunoprecipitation experiments were conducted with FLAG-tagged CCM2 and GFP-tagged KRIT1. Experiments were performed with wild-type CCM2 and either WT KRIT1 or mutants within its NPX(Y/F) motifs (KRIT1NPX(Y/F)1 mutant, Y195A; KRIT1NPX(Y/F)2 mutant, F234A; KRIT1NPX(Y/F)3 mutants, Y252A/F253A; KRIT1NPX(Y/F)2 and KRIT1NPX(Y/F)3 mutants, F234A/Y252A/F253A) using GFP only as a negative control. C, representative pull-down assay. GST fusion KRIT1 NPX(Y/F) motifs were immobilized on beads and incubated with purified CCM2PTB. Coomassie staining is shown. D, quantification of pull-down experiments (n = 5) expressed as a percentage of wild-type CCM2PTB binding within each experiment and averaged across experiments. Error bars, S.E.; ****, p < 0.0001. E, size exclusion chromatography with multiangle light scattering analysis of CCM2PTB-KRIT1NPX(Y/F)3 complex shows that it elutes as a peak of molar mass 20.8 kDa ± 1.1%.
