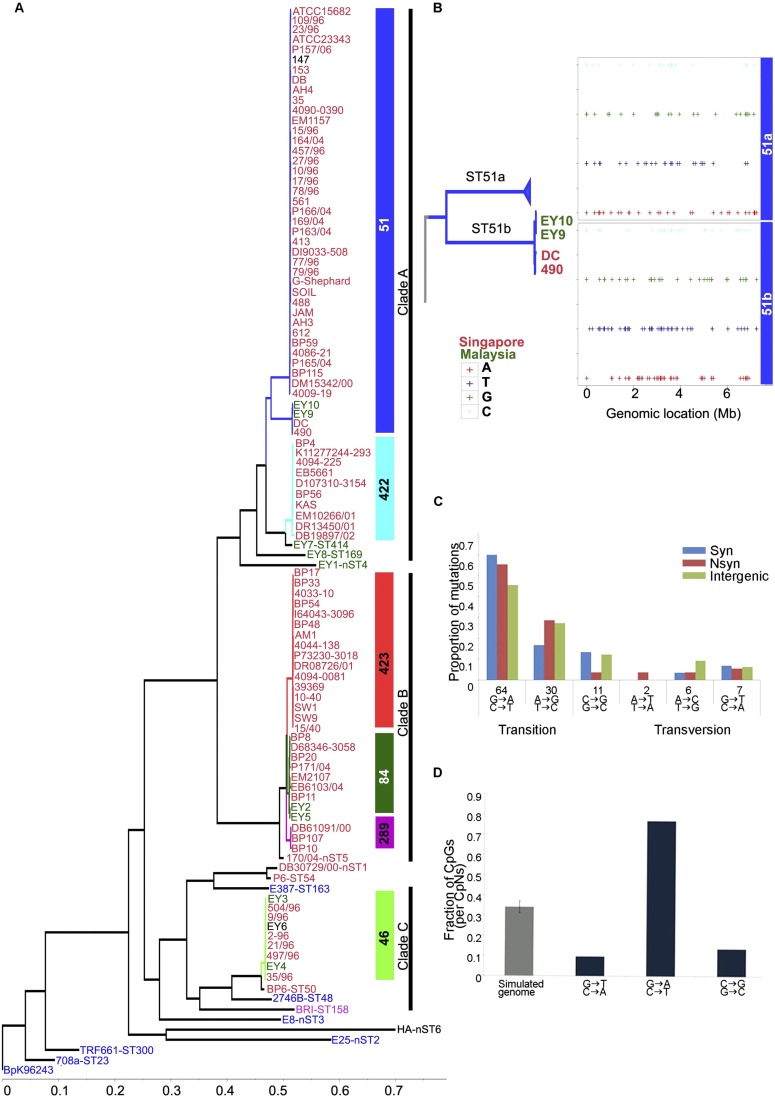
Figure 1.
Whole-genome phylogeny and sequence variation of Bp strains. (A) Global phylogeny of Bp strains. The maximum likelihood tree was constructed using SNPs not associated with recombination events (see Results). Tip labels are colored according to the geographic locations of isolation ([red] Singapore; [green] Malaysia; [blue] Thailand; [black] unknown; [pink] imported to UK). Inset bars at right indicate the MLST scheme ([blue] ST51; [cyan] ST422; [red] ST423; [dark green] ST84; [pink] ST289; [light green] ST46). Three major genomic clades are identified: Clade A (ST51, ST422, ST414, ST169, nST4); Clade B (ST423, ST84, ST289, nST5); and Clade C (ST46, ST50). (B) Intra-ST subgroups resolved by WGS. ST51 strains cluster into two groups: ST51a and ST51b. Genomic locations of 342 L-SNPs (including both intra- and intergenic SNPs) distinguishing ST51a and ST51b are shown. The top and bottom panels with four rows show SNPs exclusively present in the two groups ST51a+ST51b− and ST51a−ST51b+, respectively. (C) Mutation spectra of ST51a: relative rates of six possible mutation categories. The most common mutations are C/G → T/A transitions. (D) Fraction of the three classes of cytosine mutations occurring at CG dinucleotides in the Bp genome compared with the expected fraction based on the average of 100 simulated genomes of the same size and composition (gray).