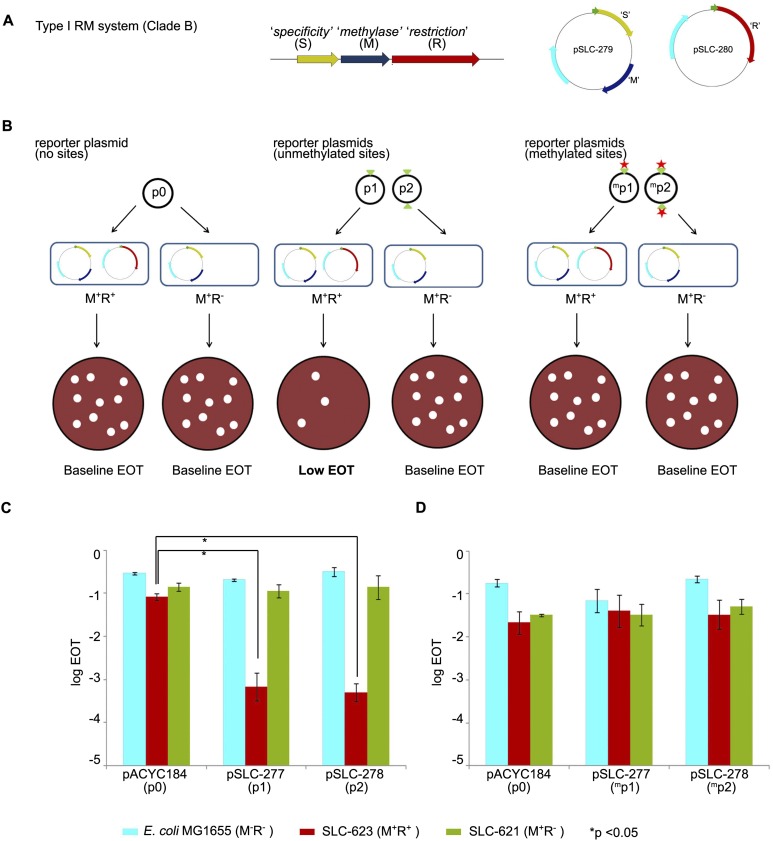
Figure 5.
Restriction of non-self DNA by clade-specific Bp RM systems. (A) Molecular cloning of a Type I RM system specific to Bp genomic Clade A. The RM system comprises three genes: (R) “restriction,” (M) “methylase,” (S) “specificity.” Genes S (yellow) and M (blue) were cloned in plasmid pSLC-279 with kanamycin resistance (KmR) to give the M+ plasmid. Gene R (red) was cloned in plasmid pSLC-280 with ampicillin resistance (ApR) to give the R+ plasmid. Resistance genes are depicted in cyan. Green arrows represent the T5 promoter used to induce expression of the cloned genes. Plasmids are not drawn to scale. (B) Efficiency of transformation (EOT) assay. Reporter plasmids p0, p1, and p2 harbor zero, one, and two copies of the predicted Type I recognition site (5′-GTCATN5TGG-3′; indicated by green triangles). Plasmid p0 should not show any EOT changes because it does not contain Type I recognition sequences. Unmethylated plasmids p1 and p2, when transformed into M+R+ strains, should be recognized via their Type I sites and cleaved by the Type I restriction enzyme (registered as a drop in number of transformants). However, when transformed into M+R− strains that express the methyltransferase alone, no EOT differences should be observed. In contrast, methylated p1 and p2 plasmids (obtained by passage through M+R− strains; methylated sites indicated by red stars and superscript m), when transformed into M+R+ strains, should be recognized as “self” DNA by the Type I system and resist cleavage, resulting in minimal EOT changes. (C) EOT assay results using unmethylated plasmids. Host strains are MG1655 (M−R−, no RM system, cyan); SLC-623 (M+R+, complete RM system, red); and SLC-621 (M+R−, methyltransferase only, green). Reporter plasmids are pACYC184 (p0, control plasmid); pSLC-277 (p1, 1 recognition site); and pSLC-278 (p2, 2 recognition sites). Significant differences in EOT are observed between control plasmid p0 and plasmids p1 and p2 when transformed into M+R+ strains (P < 0.01) but not in host E. coli or M+R− strains. EOT in this study is the normalized number of CmR transformants obtained per unit amount of plasmid DNA. (D) EOT assay using methylated plasmids. Reporter plasmids were passaged through M+R− strains prior to transformation, which is predicted to cause recognition site methylation. No significant EOT differences are observed across the strains. All experiments were performed in triplicate, and data are presented as mean and standard deviations. Data are presented as log10 values of EOT. Student’s t-test was used to test for significant differences.