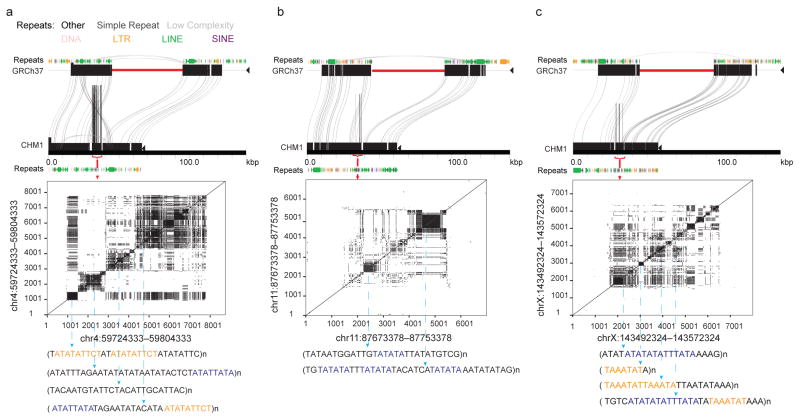
Extended Data Figure 1. Sequence content of gap closures.
a, Gap closures are enriched for simple repeats compared to equivalently sized regions randomly sampled from GRCh37; examples of the organization of these regions is shown using Miropeats for (b) chromosome 4 (GRCh37, chr4:59724333-59804333), (c) chromosome 11 (GRCh37, chr11:87673378-87753378), and (d) chromosome X (GRCh37, chrX:143492324-143572324). Dotplots show the architecture of the degenerate STRs with the core motif highlighted below. Shared sequence motifs between blocks is indicated by color.