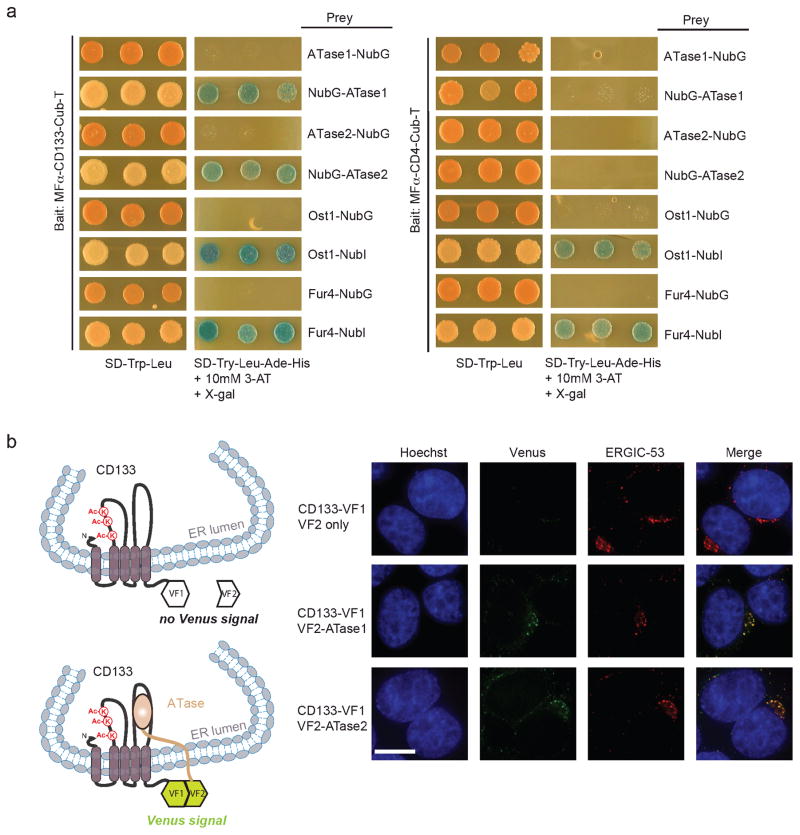
Figure 3. ATase1 and ATase2 physically interact with CD133 in the ERGIC compartment.
a) Membrane yeast two-hybrid (MYTH) was performed as previously described (4, 8). Generation of NubG fused ATase1 or ATase2 was performed by PCR amplification from pDONR223-NAT8 and pDONR223-NAT8B (Open Biosystems Inc.), respectively, with the following primers for N’terminal tagged NubG: sense 5′-GGT GGT CCA TAC CCA TAC GAT GTT CCA GAT TAC GCT GCT CCT TGT CAC ATC CGC AAA TAC-3′ and antisense 5′-GTA AGC GTG ACA TAA CTA ATT ACA TGA CTC GAG TCA CAG ACT CCC TAC C-3′ and C’terminal tagged NubG: sense 5′ CAA TAT TTC AAG CTA TAC CAA GCA TAC AAT CAA CTC AAT GGC TCC TTG TCA CAT CCG-3′ and antisense 5′-GGA GCG TAA TCT GGA ACA TCG TAT GGG TAC ATA TCC AGA CTC CCT ACC TTA GAA G-3′. PCR products were introduced into the pPR3-N or pPR3-C MYTH prey vectors by yeast homologous recombination as previously described (8). MFα-CD133-Cub-T or the control MFα-CD4-Cub-T were expressed in the yeast strain THY.AP4 along with NubG-ATase1, NubG-ATase2, ATase1-NubG or ATase2-NubG and assayed for growth on selective media. MFα-CD133-Cub-T and MFα-CD4-Cub-T were also expressed in THY.AP4 along with positive controls Ost1-NubI and Fur4-NubI and the negative controls Ost1-NubG and Fur4-NubG to demonstrate absence of self-activation and proper membrane integration. b) CD133 (pDONR223-CD133) (3), ATase1 (pDONR223-NAT8B) and ATase2 (pDONR223-NAT8) (Open Biosystems Inc.) were introduced into Gateway-compatible protein complementation assay (PCA) vectors. Vectors were transfected into HEK293 cells and immunofluorescence was performed as previously described (4), using the anti-ERGIC-53 antibody (C-6, Santa Cruz Biotechnology Inc.). CD133-VF1 and either VF2 only, VF2-ATase1 or VF2-ATase2 were co-transfected into HEK293 cells. 48 hours post-transfection, immunofluorescence was performed to determine co-localization between ERGIC-53, CD133-VF1 and either VF2 only, VF2-ATase1 or VF2-ATase2. Cells were stained with Hoechst 33342 to visualize nuclei. Scale bar, 10μm.