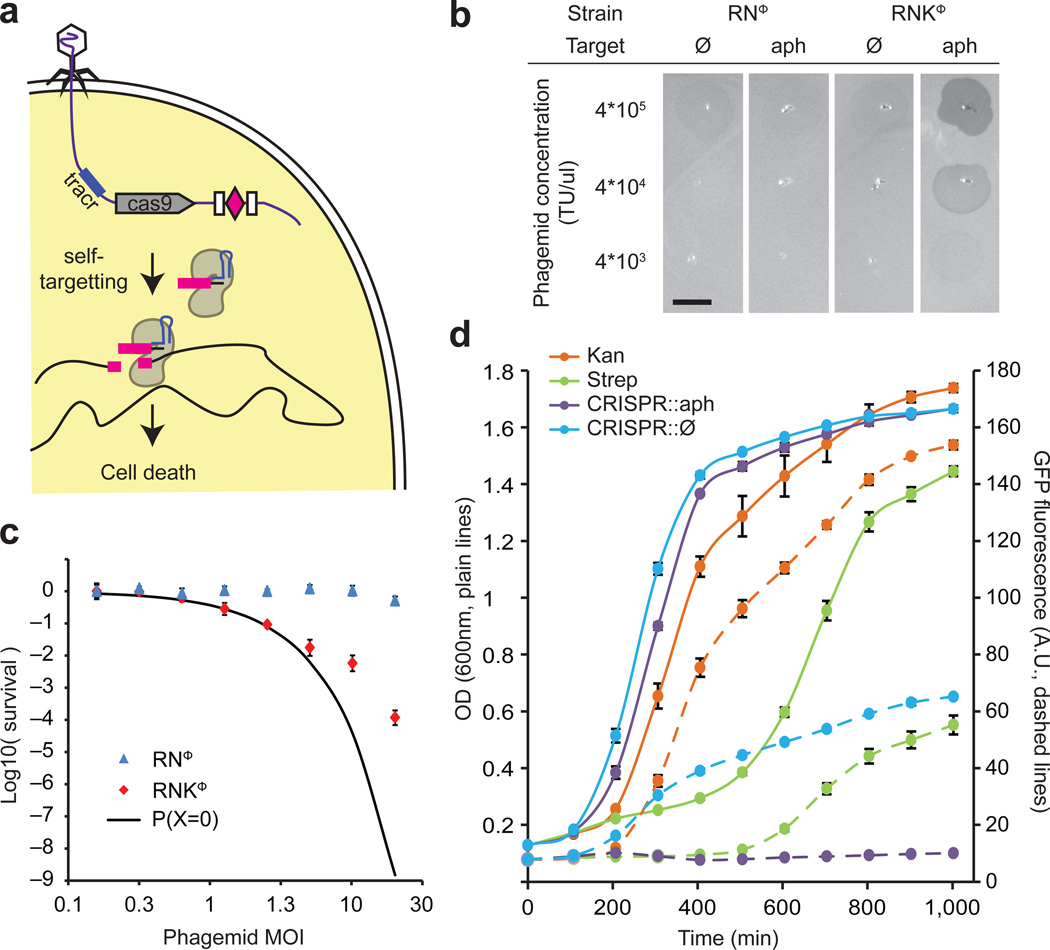
Figure 1.
Sequence-specific killing of S. aureus by a phagemid-delivered CRISPR system. (a) The ΦNM1 phage delivers the pDB121 phagemid to S. aureus cells. pDB121 carries the S. pyogenes tracrRNA, cas9 and a programmable CRISPR array sequence. Expression of cas9 and a self-targeting crRNA leads to chromosome cleavage and cell death. (b) Lysates of pDB121 phagemid targeting the aph-3 kanamycin resistance gene or a non-targeting control are spotted on top-agar lawns of either RNΦ or RNKΦ cells. Scale bar, 5mm (c) Treatment of RNΦ (blue triangles) or RNKΦ (red diamonds) with pDB121::aph at various MOI. Survival is calculated as the ratio of CFU recovered after treatment to CFU from an untreated sample of the same culture (mean ± s.d.). The black curve represents the probability that a cell does not receive a phagemid making the assumption that all cells have the same chance of receiving phagemid. (d) Time course of treatment of RNKΦ/pCN57 (GFP reporter plasmid) cells in a mixed culture with non-targeted RNΦ cells. Plain lines show OD and dashed lines GFP. Kanamycin (25 ug/ml) is shown in orange, streptomycin (10 ug/ml) in green, pDB121::aph (MOI ~20) in purple, pDB121::Ø (MOI ~20) in blue.