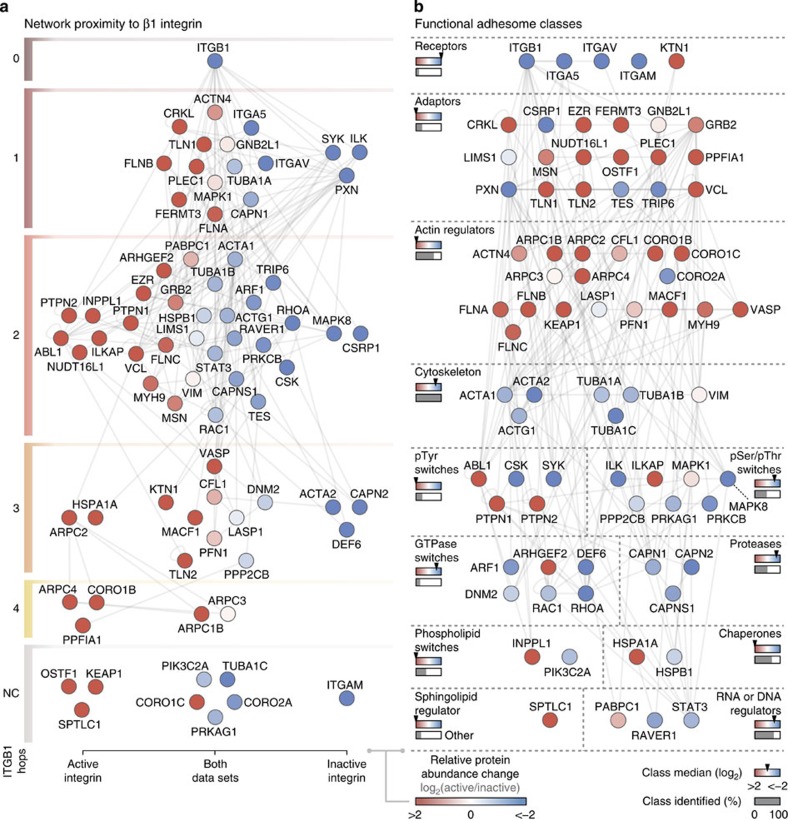
Figure 2. Analysis of the activation state-dependent adhesome network.
(a) The interaction network of adhesome components4 identified by MS was arranged according to the number of reported protein interactions (hops) from β1 integrin (ITGB1). Proteins were clustered by their detection in active (left), inactive (right) or both (middle) integrin data sets. NC, not connected. (b) The identified adhesome network was arranged according to functional class3,4. Arrowheads on coloured bars indicate median protein enrichment for each class; grey bars indicate proportion of reported adhesome class identified by MS. Sphingolipid regulator quantification was derived from ‘other’ adhesome class. ‘Channel’ and ‘E3 ligase’ adhesome classes were not represented (0% identified). Nodes (proteins) are coloured according to their enrichment in active (red) or inactive (blue) integrin complexes (log2 transformed). Gene symbols are shown for clarity (see Supplementary Table 1 for protein names).