Figure 1.
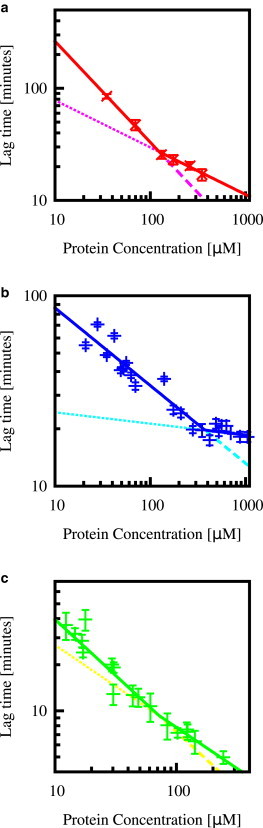
Lag time as a function of protein concentration for (a) bovine insulin, (b) lysozyme, and (c) β2 microglobulin, in all cases averaged over replicate experiments. The data in panel c was kindly provided by Xue et al. (9). The error bars indicate the standard deviation among replicate experiments at a given concentration, and the lines indicate the best fit of the power law τlag ∝ mtot−γ to data for a specified range of protein concentrations. (a) Two data sets for bovine insulin: (black) data fitted by the single power law over all protein concentrations (resulting in γ = 0.36) indicate experiments in the absence of NaCl; (red) data fitted by two power laws indicate self-assembly in the presence of 0.49 M NaCl. Each point corresponds to the mean and standard deviation of 140–200 kinetic traces at each of six protein concentrations (see Materials and Methods). Fitting the data for bovine insulin in the presence of NaCl in the range mtot ≤ 160 μM results in γ = 0.90(2) (dashed line). Fitting the data in the range mtot ≥ 160 μM results in γ = 0.42(10) (dotted line). (b) Data for lysozyme (for conditions, see Materials and Methods), where each point corresponds to the mean and standard deviation of 12–60 kinetic traces at each of 25 protein concentrations (see Materials and Methods). Fitting this data in the range mtot ≤ 300 μM results in γ = 0.41(6) (dashed line); while fitting the data in the range mtot ≥ 300 μM results in γ = 0.06(5) (dotted line). (c) Data for β2 microglobulin, where each point corresponds to the mean and standard deviation of 235 kinetic traces at 20 different protein concentrations (for conditions, see Materials and Methods and Xue et al. (9)). Fitting the data in the range mtot ≥ 30 μM gives γ = 0.77(14) (dashed line), while fitting the data in the range mtot ≥ 40 μM gives γ = 0.54(5) (dotted line). To see this figure in color, go online.
