Figure 2.
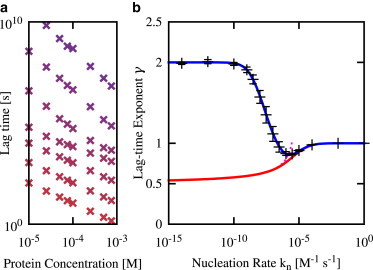
Lag-time scaling with protein concentration for the fragmentation model depends on the primary nucleation rate constant. (a) Lag-time versus concentration curves obtained from stochastic simulations, for several values of the nucleation rate constant kn: kn increases top to bottom in factors of 100, from 10−12 M−1 s−1 (purple) to 1 M−1 s−1 (red). (b) Lag-time scaling exponent γ as a function of primary nucleation rate constant kn. (Black symbols) Simulation results obtained by fitting the data shown in panel a to log(τlag) = log(A) – γlog(mtot). The error bars are dominated by the confidence in the fit, rather than the error in the lag-time measurements. (Red line) The value γ(kn) extracted from the full prediction of the fragmentation model, Eq. 7. (Magenta line) The value γ(kn) extracted from the approximate prediction of the fragmentation model, Eq. 8. (Blue line) The value γ(kn) extracted from the lag-time expression Eq. 9—i.e., the full fragmentation model solution augmented by an extra term to describe the first primary nucleation event. For all the data shown, the protein concentration range was 1 × 10−5 ≤ mtot ≤ 7.5 × 10−4 M and the parameters were nc = 2, k+ = 5 × 104 M−1 s−1, kf = 3 × 10−8 s−1, and the volume V = 0.83 pL. To see this figure in color, go online.
