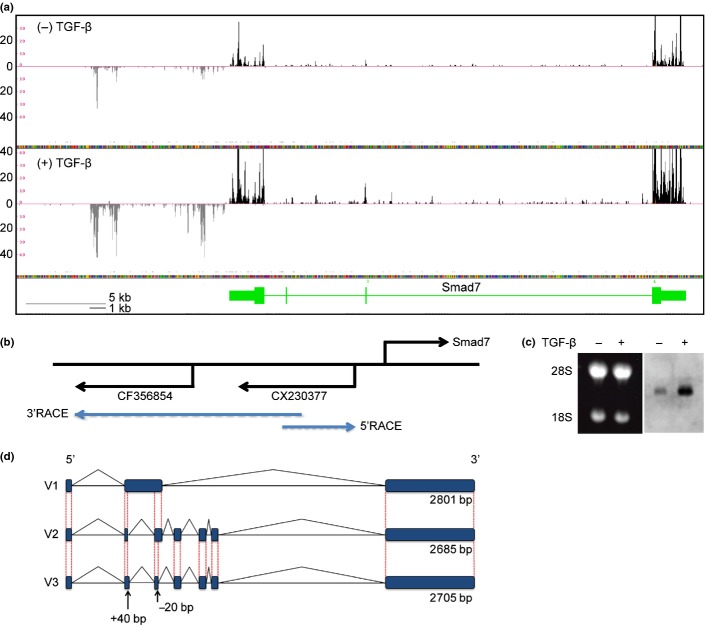
Fig. 1.
Cloning of lncRNA-Smad7. (a) RNA-sequencing data from NMuMG cells at the mouse Smad7 gene locus. Black and grey tags are aligned to the sense and antisense genomic sequences, respectively. Vertical axis: mapped tag numbers. Smad7 gene is shown in green, and its exons are shown as boxes (thin boxes, untranslated regions; thick boxes, coding regions) connected by horizontal lines representing introns. (b) Relative positions of the Smad7 gene and EST clones with direction of transcription. Positions of the gene-specific primers used for RACE are shown at the back ends of the blue arrows. (c) Northern blotting of lncRNA-Smad7. NMuMG cells were treated with TGF-β or left untreated for 24 h. Left panel: Ethidium bromide staining of the gel to show the positions of 28s/18s rRNA and equal loading of the samples. Right panel: Northern blotting with an EST sequence (CX230377) as a probe. (d) Schematic representation of the identified lncRNA-Smad7 transcripts. V3 transcript was identified by quantitative RT-PCR (qRT-PCR) analysis of JygMC(A) and confirmed by sequencing of the PCR product. V3 had a longer exon 2 and a shorter exon 3 than V2 transcript.