Abstract
The epithelial cell adhesion molecule (EpCAM) is overexpressed in a wide variety of human cancers and is associated with patient prognosis, including those with lung cancer. However, the association of single nucleotide polymorphisms (SNPs) in the EpCAM gene with the prognosis for non-small-cell lung cancer (NSCLC) patients has never been investigated. We evaluated the association between two SNPs, rs1126497 and rs1421, in the EpCAM gene and clinical outcomes in a Chinese cohort of 506 NSCLC patients. The SNPs were genotyped using the Sequenom iPLEX genotyping system. Multivariate Cox proportional hazards model and Kaplan–Meier curves were used to assess the association of EpCAM gene genotypes with the prognosis of NSCLC. We found that the non-synonymous SNP rs1126497 was significantly associated with survival. Compared with the CC genotype, the CT+TT genotype was a risk factor for both death (hazard ratio, 1.40; 95% confidence interval [CI], 1.02–1.94; P = 0.040) and recurrence (hazard ratio, 1.34; 95% CI, 1.02–1.77; P = 0.039). However, the SNP rs1421 did not show any significant effect on patient prognosis. Instead, the AG+GG genotype in rs1421 was significantly associated with early T stages (T1/T2) when compared with the AA genotype (odds ratio for late stage = 0.65; 95% CI, 0.44–0.96, P = 0.029). Further stratified analysis showed notable modulating effects of clinical characteristics on the associations between variant genotypes of rs1126497 and NSCLC outcomes. In conclusion, our study indicated that the non-synonymous SNP rs1126497 may be a potential prognostic marker for NSCLC patients.
Keywords: Chinese population, EpCAM, non-small-cell lung cancer, prognosis, single nucleotide polymorphism
Lung cancer is the most common malignancy in the world.1 It is a main cause of cancer-associated death in both men and women. Non-small-cell lung cancer (NSCLC) accounts for approximately 80% of primary lung cancers.2 Tobacco use is known as the most significant risk factor leading to lung cancer. Other known risk factors for lung cancer include environmental and occupational exposures. However, the major causes underlying lung cancer development are genetic and epigenetic damage resulting from environmental carcinogens.3 In spite of advances achieved in diagnosis and treatment in recent years, the clinical outcome for patients with lung cancer remains disappointing. The 5-year survival rate of NSCLC patients who receive surgical resection is only approximately 15%.4 Although the key prognostic determinant in lung cancer is clinical stage at diagnosis, it is common to observe that patients at a similar stage have considerably different clinical outcomes. In fact, it is beneficial to improve survival rates after surgical operations by finding suitable biomarkers for prognosis prediction in lung cancer.5
The EpCAM gene is located on chromosome 2q21, which encodes a carcinoma-associated type I membrane protein of 314 amino acids.6 It is expressed in a variety of human epithelial tissues and cancer tissues, as well as progenitor and stem cells.7 Previous studies have reported that epithelial cell adhesion molecule (EpCAM) plays diverse roles in different cancers.8–11 Several studies have shown that EpCAM is overexpressed in many different types of cancers, such as NSCLC,12 hepatocellular carcinoma,13 and prostate cancer.14 Overexpressed EpCAM has been confirmed to increase the expression of c-Myc and cyclins A and E, and then induce a direct impact on cell cycle control.15 Cells with high levels of EpCAM have a reduced requirement for growth factors; they grow in an anchorage-independent manner and proliferate more rapidly. However, previous studies have also reported that EpCAM has protective effects in some cancers such as renal cell, thyroid, oral squamous cell, and gastric carcinomas.16–18 Therefore, whether EpCAM functions as an oncogene or as a tumor suppressor gene mainly relies on the tumor microenvironment.8 With regard to lung cancer, cumulative experimental evidence has shown that knockdown of EpCAM inhibits tumor growth and results in massive apoptosis of cancer cells, which indicates a promising therapeutic target for lung cancer patients.19
Single nucleotide polymorphisms located in encoded regions that alter the amino acid sequence of a protein are termed non-synonymous SNPs (ns-SNPs). Recently, ns-SNPs have been investigated as biomarkers for risk assessment and prognosis prediction, as they are closely associated with a variety of diseases and their development.20,21 Previously published reports showed that an ns-SNP in the EpCAM gene (rs1126497) is associated with risk of breast cancer and cervical cancer.22,23 In addition, EpCAM plays an important role in hereditary diseases of non-polyposis colorectal cancer (Lynch syndrome) and congenital tufting enteropathy.24 Another study reported that a nonsense mutation in the EpCAM gene is significantly associated with congenital tufting enteropathy patients.25 All of these results show considerable functions of the EpCAM gene in human hereditary and tumor diseases.
In this study, we selected two SNPs in the EpCAM gene and assessed their effects on the clinical outcomes of NSCLC patients. To the best of our knowledge, this is the first study on the prognostic role of SNPs in the EpCAM gene in NSCLC.
Materials and Methods
Study group
A total of 577 patients with NSCLC who had received surgical resection treatment were initially recruited into an ongoing molecular epidemiological study at the Department of Thoracic Surgery at Tangdu Hospital (Fourth Military Medical University, Xi'an, China) between July 2009 and February 2012. None of the patients had been treated with surgery, chemotherapy, and/or radiotherapy before enrolment into the study. There was no restriction for age, gender, or disease stage at enrolment, but the study was restricted to patients with the NSCLC type of lung cancer. In this prognosis study, we excluded 71 patients, including 32 who had incomplete clinical information or failed follow-up, 13 who died within 2 months after surgery, 17 patients with metastatic diseases, and nine with recurrence within 1 month after surgery. Finally, 506 NSCLC patients were included in the present study and successfully genotyped. All patients were Han Chinese. This study was approved by the Institutional Review Boards of the Fourth Military Medical University, and signed informed consent was obtained from each participant.
Demographic and clinical data
Demographic data was collected through in-person interviews using a standard epidemiological questionnaire, including age, gender, ethnicity, residential region, smoking status, alcohol use, education status, and family history of cancer. Detailed clinical information was collected through medical chart review, including medical imaging data, pathological diagnosis, TNM stage, and treatment information. A standard follow-up was carried out by a trained clinical specialist through medical record review or telephone interview. The latest follow-up data in this analysis were obtained in November 2012, and 41 (8.1%) patients were lost during follow-up and censored for the analyses. All the patients donated 5 mL blood before treatment for genomic DNA extraction using the EZNA Blood DNA Midi Kit (Omega Bio-Tek, Norcross, GA, USA) in the laboratory.
Single nucleotide polymorphism selection and genotyping
Functional SNPs in the EpCAM gene were selected using a set of Web-based SNP selection tools. We found two functional SNPs, rs1421 and rs1126497. The former, s1421, is located in the 3′-UTR, which was predicted to be a potential microRNA binding site by the FuncPred tool (available from http://snpinfo.niehs.nih.gov/snpinfo/snpfunc.htm), and may influence the mRNA degradation or translation. The SNP rs1126497 is an ns-SNP located in exon 3, which changes the 115 Met to Thr and may affect the protein function. Genotyping was carried out using the iPLEX genotyping system (Sequenom, San Diego, CA, USA). Laboratory personnel who undertook the genotyping assays were blinded to patients' information. The average call rate for the genotyping was 98.5%. Strict quality control measures were implemented during genotyping with more than 99% concordance in samples that were randomly selected to be genotyped in duplicate.
Statistical analysis
The spss version 19.0 statistical software (SPSS Inc., Chicago, IL, USA) was used for all statistical analyses. Three genetic models (additive, dominant, and recessive) were applied to assess the association between single SNP and clinical outcomes of NSCLC patients. Two major endpoints, overall survival (OS) and recurrence-free survival (RFS), were analyzed in this study. Overall survival was defined as the time from diagnosis to death from any cause; RFS was defined as the time from diagnosis to the first date when patients developed recurrence. Considering that there was a very small number (≤5) of patients with the rare homozygous variant genotype for these two SNPs of the EpCAM gene, we did not present the data from the recessive model in our analyses to avoid unstable estimations. Hazard ratios (HRs) and 95% confidence intervals (CIs) were estimated from a multivariate Cox proportional hazards model, adjusting for age, gender, smoking status, histology, TNM stage, differentiation, and adjuvant chemotherapy or radiotherapy. Kaplan–Meier curves and the log–rank test were used to assess the differences among patient groups in OS and RFS analyses. All P-values in this study were two-sided. P < 0.05 was considered statistically significant.
Results
Demographic and clinical characteristics and prognosis analysis
A total of 506 NSCLC patients were included in this study from Tangdu Hospital. They were an ethnically homogenous group of Han Chinese, the median age was 59 years (range, 27–86 years), and 392 (77.5%) were male patients. Only 160 (31.6%) patients were never smokers, and all others had a smoking history. Of the total 506 patients, 271 were diagnosed with squamous cell carcinoma, 152 with adenocarcinoma, 62 with adenosquamous carcinoma, 16 with carcinosarcoma, three with large-cell lung cancer, and two with mucoepidermoid carcinoma. There were 284 (56.1%) patients diagnosed with stage I and stage II disease, and 222 (43.9%) patients had stage III disease. There were 347 (68.6%) patients with well or moderately differentiated tumors, and 159 (31.4%) with poorly differentiated or undifferentiated tumors. According to the records, 375 patients received adjuvant chemotherapy or radiotherapy.
Associations between patients' characteristics and NSCLC outcomes
We carried out a multivariate Cox regression analysis to assess the associations between clinical characteristics and outcomes in NSCLC patients. As shown in Table1, patients with adenocarcinoma showed better OS (HR = 0.61; 95% CI, 0.39–0.93) than those with squamous cell carcinoma. Patients at advanced tumor stage had significantly increased risk for death and recurrence with HRs of 1.94 (95% CI, 1.41–2.68) and 2.01 (95% CI, 1.53–2.65), respectively. Patients with poorly differentiated or undifferentiated tumors showed significant association with worse RFS (HR for recurrence = 1.61; 95% CI, 1.14–2.28). In addition, adjuvant chemotherapy or radiotherapy significantly improved the OS of patients with NSCLC (HR for death = 0.67; 95% CI, 0.47–0.69).
Table 1.
Distribution of characteristics in Chinese patients with non-small-cell lung cancer (n = 506) and prognosis analysis
| Variables | No. of total patients |
Overall survival |
Recurrence-free survival |
||||
|---|---|---|---|---|---|---|---|
| Deaths | HR (95% CI)† | P-value | Recurrence | HR (95% CI)† | P-value | ||
| n = 506, no. (%) | n = 159, no. (%) | n = 220, no. (%) | |||||
| Age | |||||||
| ≤59 years | 251 (49.6) | 77 (48.4) | Ref. | 117 (53.2) | Ref. | ||
| >59 years | 255 (50.4) | 82 (51.6) | 0.99 (0.72–1.38) | 0.974 | 103 (46.8) | 0.87 (0.66–1.15) | 0.340 |
| Gender | |||||||
| Female | 114 (22.5) | 36 (22.6) | Ref. | 54 (24.5) | Ref. | ||
| Male | 392 (77.5) | 123 (77.4) | 0.97 (0.57–1.66) | 0.917 | 166 (75.5) | 1.12 (0.71–1.78) | 0.622 |
| Smoking status | |||||||
| Never smoker | 160 (31.6) | 50 (31.4) | Ref. | 75 (34.1) | Ref. | ||
| Ever smoker | 346 (68.4) | 109 (68.6) | 0.97 (0.59–1.58) | 0.894 | 145 (65.9) | 0.78 (0.51–1.17) | 0.231 |
| Histology | |||||||
| Squamous cell carcinoma | 271 (53.6) | 82 (51.6) | Ref. | 112 (50.9) | Ref. | ||
| Adenocarcinoma | 152 (30.0) | 35 (22.0) | 0.61 (0.39–0.93) | 0.022 | 59 (26.8) | 0.77 (0.54–1.09) | 0.135 |
| Others‡ | 83 (16.4) | 42 (26.4) | 1.35 (0.83–2.22) | 0.230 | 49 (22.3) | 1.11 (0.72–1.71) | 0.643 |
| TNM stage | |||||||
| I/II | 284 (56.1) | 72 (45.3) | Ref. | 98 (44.5) | Ref. | ||
| III | 222 (43.9) | 87 (54.7) | 1.94 (1.41–2.68) | <0.001 | 122 (55.5) | 2.01 (1.53–2.65) | <0.001 |
| Differentiation | |||||||
| Well/moderate | 347 (68.6) | 88 (55.3) | Ref. | 129 (58.6) | Ref. | ||
| Poorly/undifferentiated | 159 (31.4) | 71 (44.7) | 1.50 (0.99–2.26) | 0.053 | 91 (41.4) | 1.61 (1.14–2.28) | 0.007 |
| Adjuvant chemotherapy or radiotherapy | |||||||
| No | 131 (25.9) | 46 (28.9) | Ref. | 54 (24.5) | Ref. | ||
| Yes | 375 (74.1) | 113 (71.1) | 0.67 (0.47–0.96) | 0.030 | 166 (75.5) | 0.86 (0.62–1.19) | 0.365 |
Adjusted by age, gender, smoking status, histology, TNM stage, differentiation, and adjuvant chemotherapy or radiotherapy where appropriate.
Other carcinomas include adenosquamous carcinoma, large cell carcinoma, carcinosarcoma, and mucoepidermoid carcinoma. CI, confidence interval; HR, hazard ratio; Ref., reference.
Associations between SNPs and clinical outcome of NSCLC
The Multivariate Cox regression model was used to evaluate the effect of EpCAM SNPs on death and recurrence in NSCLC patients. As shown in Table2, patients carrying CT or TT genotypes in the SNP rs1126497 showed a significantly higher death risk when compared with those carrying the CC genotype (HR for additive model = 1.46; 95% CI, 1.09–1.97, P = 0.012; HR for dominant model = 1.41; 95% CI, 1.02–1.94, P = 0.039). Similar results were observed in the RFS analysis (HR for additive model = 1.37; 95% CI, 1.06–1.76, P = 0.016; HR for dominant model = 1.34; 95% CI, 1.01–1.77, P = 0.039). Additionally, there were no significant results observed for the SNP rs1421 in either the OS or RFS analyses. For rs1126497, Kaplan–Meier curves significantly distinguished the patient groups with different genotypes in the OS analysis (log–rank P for additive and dominant models = 0.007 and 0.021, respectively) and RFS analysis (log–rank P for additive and dominant models = 0.008 and 0.006, respectively) (Fig.1).
Table 2.
Association between single nucleotide polymorphisms (SNPs) in the EpCAM gene and clinical outcomes in Chinese patients with non-small-cell lung cancer
| SNP locus | Genotype | Overall survival |
Recurrence-free survival |
||||
|---|---|---|---|---|---|---|---|
| Death/total | HR (95% CI)† | P-value | Recurrence/total | HR (95% CI)† | P-value | ||
| rs1421 miRNA binding site | AA | 108/329 | Ref. | 144/329 | Ref. | ||
| AG | 44/147 | 1.14 (0.79–1.63) | 0.485 | 67/147 | 1.31 (0.97–1.77) | 0.078 | |
| GG | 6/18 | 1.33 (0.57–3.07) | 0.508 | 7/18 | 0.81 (0.38–1.75) | 0.596 | |
| Dominant | 50/165 | 1.16 (0.82–1.64) | 0.404 | 74/165 | 1.24 (0.93–1.65) | 0.151 | |
| Additive | 1.14 (0.86–1.53) | 0.364 | 1.12 (0.88–1.41) | 0.360 | |||
| rs1126497 non- synonymous SNP | CC | 93/336 | Ref. | 131/336 | Ref. | ||
| CT | 59/157 | 1.34 (0.96–186) | 0.087 | 80/157 | 1.29 (0.97–1.71) | 0.076 | |
| TT | 5/10 | 3.44 (1.37–8.65) | 0.006 | 6/10 | 2.61 (1.14–6.02) | 0.024 | |
| Dominant | 64/167 | 1.40 (1.02–1.94) | 0.040 | 86/167 | 1.34 (1.02–1.77) | 0.039 | |
| Additive | 1.46 (1.08–1.96) | 0.013 | 1.37 (1.06–1.76) | 0.016 | |||
Adjusted by age, gender, smoking status, histology, TNM stage, differentiation, and adjuvant chemotherapy or radiotherapy. CI, confidence interval; HR, hazard ratio; Ref., reference.
Figure 1.
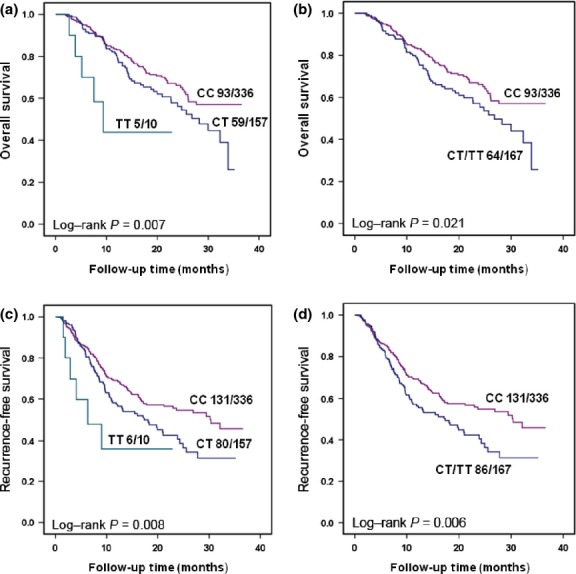
Kaplan–Meier plots of non-small-cell lung cancer (NSCLC) patients with different genotypes of rs1126497 in the EpCAM gene. (a) Overall survival of NSCLC patients by rs1126497 using additive genetic model. (b) Overall survival of NSCLC patients by rs1126497 using dominant genetic model. (c) Recurrence-free survival of NSCLC patients by rs1126497 using additive genetic model. (d) Recurrence-free survival of NSCLC patients by rs1126497 using dominant genetic model.
Associations between EpCAM polymorphisms and NSCLC progression
We applied binary logistic regression to explore the association between EpCAM SNPs and cancer progression. As shown in Table3, the AG+GG genotype in rs1421 was significantly associated with early T stages (T1/T2) when compared with the AA genotype (odds ratio [OR] for late stage = 0.65; 95% CI, 0.44–0.96, P = 0.029). However, there was no significant association between the SNP rs1421 and N stage. Additionally, the SNP rs1126497 had no significant association with either the T or N stage of NSCLC.
Table 3.
Association between single nucleotide polymorphisms (SNPs) in the EpCAM gene and disease progression in Chinese patients with non-small-cell lung cancer
| SNP | Genotype | T stage |
N stage |
||||||
|---|---|---|---|---|---|---|---|---|---|
| T1/T2 (%) | T3/T4 (%) | OR (95% CI) | P-value | N0 (%) | N1/N2/N3 (%) | OR (95% CI) | P-value | ||
| rs1421 | AA | 192 (63.0) | 137 (72.5) | Ref. | 148 (65.5) | 181 (67.5) | Ref. | ||
| AG | 101 (33.1) | 46 (24.3) | 0.64 (0.42–096) | 0.033 | 70 (31.0) | 77 (28.7) | 0.89 (0.61–1.33) | 0.594 | |
| GG | 12 (3.9) | 6 (3.2) | 0.70 (0.26–1.91) | 0.488 | 8 (3.5) | 10 (3.7) | 1.02 (0.39–2.66) | 0.964 | |
| AG+GG | 113 (37.0) | 52 (27.5) | 0.65 (0.44–0.96) | 0.029 | 78 (34.5) | 87 (32.5) | 0.91 (0.63–1.33) | 0.630 | |
| rs1126497 | CC | 215 (69.4) | 121 (62.7) | Ref. | 157 (68.0) | 179 (65.8) | Ref. | ||
| CT | 90 (29.0) | 67 (34.7) | 1.32 (0.89–1.95) | 0.156 | 72 (31.2) | 85 (31.3) | 1.04 (0.71–1.51) | 0.857 | |
| TT | 5 (1.6) | 5 (2.6) | 1.78 (0.50–6.26) | 0.371 | 2 (0.9) | 8 (2.9) | 3.51 (0.73–16.77) | 0.116 | |
| CT+TT | 95 (30.6) | 72 (37.3) | 1.35 (0.92–1.97) | 0.123 | 74 (32.0) | 93 (34.2) | 1.10 (0.76–1.60) | 0.609 | |
N stage, regional lymph node involvement; Ref., reference; T stage, size and tissue invasion of the primary tumor.
Associations between EpCAM polymorphisms and prognosis of NSCLC stratified by patient characteristics
To investigate the modulator effects of clinical characteristics on the association between EpCAM polymorphisms of rs1126497 and prognosis, we carried out multivariate Cox proportional hazard analyses in a dominant model stratified by age, gender, smoking status, histology, TNM stage, differentiation, and adjuvant chemotherapy or radiotherapy. As shown in Table4, the CT+TT genotype was significantly associated with both worse OS and RFS in NSCLC patients with older age, male gender, smoking history, squamous cell type tumor, well or moderately differentiated tumor, and adjuvant chemotherapy or radiotherapy. We observed that patients with the CT+TT genotype had significantly increased risk for death (HR = 1.60; 95% CI, 1.02–2.53) and recurrence (HR = 1.67; 95% CI, 1.11–2.51) in patients with older age. For male patients, the HRs for death and recurrence were 1.46 (95% CI, 1.02–2.10) and 1.57 (95% CI, 1.15–2.14), respectively. For ever smoker patients, the HRs for death and recurrence were 1.52 (95% CI, 1.04–2.23) and 1.73 (95% CI, 1.24–2.42), respectively. For patients with squamous cell carcinoma, the HRs for death and recurrence were 1.94 (95% CI, 1.24–3.03) and 1.54 (95% CI, 1.04–2.28), respectively. For patients with well or moderately differentiated tumors, the HRs for death and recurrence were 1.84 (95% CI, 1.20–2.81) and 1.58 (95% CI, 1.10–2.26), respectively. For patients who received adjuvant chemotherapy or radiotherapy, the HRs for death and recurrence were 1.52 (95% CI, 1.04–2.21) and 1.57 (95% CI, 1.15–2.15), respectively. Tumor stage, differentiation, and adjuvant treatments are common and important clinical variables in cancer survival analysis. Therefore, we assessed the prognostic values of rs1126497 on OS analysis stratified by these factors. As the results show in Figure2, compared to patients carrying wild-type (CC) rs1126497, patients with variant genotypes showed significantly shorter survival in the strata of patients with early-stage disease (P = 0.038), better differentiated tumors (P = 0.002), and those who received adjuvant chemotherapy or radiotherapy (P = 0.014).
Table 4.
Stratified analysis of association between single nucleotide polymorphism rs1126497 in the EpCAM gene with prognosis in Chinese patients with non-small-cell lung cancer (n = 503)
| Variables | Overall survival |
Recurrence-free survival |
||||
|---|---|---|---|---|---|---|
| Death/total |
HR (95% CI)† | Recurrence/total |
HR (95% CI)† | |||
| CC | CT/TT | CC | CT/TT | |||
| Age | ||||||
| ≤59 years | 43/160 | 32/89 | 1.15 (0.71–1.84) | 67/160 | 48/89 | 1.15 (0.78–1.69) |
| >59 years | 50/176 | 32/78 | 1.60 (1.02–2.53) | 64/176 | 38/78 | 1.67 (1.11–2.51) |
| Gender | ||||||
| Female | 23/80 | 12/32 | 1.00 (0.46–2.19) | 37/80 | 15/32 | 0.58 (0.29–1.17) |
| Male | 70/256 | 52/135 | 1.46 (1.02–2.10) | 94/256 | 71/135 | 1.57 (1.15–2.14) |
| Smoking status | ||||||
| Never | 32/114 | 17/45 | 0.82 (0.42–1.60) | 51/114 | 23/45 | 0.60 (0.34–1.07) |
| Ever | 61/222 | 47/122 | 1.52 (1.04–2.23) | 80/222 | 63/122 | 1.73 (1.24–2.42) |
| Histology | ||||||
| Squamous cell carcinoma | 41/175 | 40/94 | 1.94 (1.24–3.03) | 61/175 | 49/94 | 1.54 (1.04–2.28) |
| Adenocarcinoma | 23/106 | 11/45 | 0.78 (0.37–1.65) | 38/106 | 20/45 | 1.04 (0.59–1.82) |
| Others‡ | 29/55 | 13/28 | 1.01 (0.50–2.04) | 32/55 | 17/28 | 1.18 (0.63–2.21) |
| TNM stage | ||||||
| I/II | 43/196 | 29/87 | 1.62 (0.99–2.62) | 61/196 | 36/87 | 1.45 (0.96–2.20) |
| III | 50/140 | 35/80 | 1.27 (0.81–1.99) | 70/140 | 50/80 | 1.26 (0.86–1.86) |
| Differentiation | ||||||
| Well/moderate | 47/230 | 40/115 | 1.84 (1.20–2.81) | 74/230 | 53/115 | 1.58 (1.10–2.26) |
| Poorly/undifferentiated | 46/106 | 24/52 | 0.93 (0.55–1.55) | 57/106 | 33/52 | 1.08 (0.69–1.69) |
| Adjuvant chemotherapy or radiotherapy | ||||||
| No | 30/91 | 15/38 | 1.12 (0.57–2.21) | 36/91 | 16/38 | 0.69 (0.37–1.33) |
| Yes | 63/245 | 49/129 | 1.52 (1.04–2.21) | 95/245 | 70/129 | 1.57 (1.15–2.15) |
Adjusted for age, gender, smoking status, histology, TNM stage, differentiation, adjuvant chemotherapy or radiotherapy, where appropriate.
Other carcinomas include adenosquamous carcinoma, large cell carcinoma, carcinosarcoma, and mucoepidermoid carcinoma. CI, confidence interval; HR, hazard ratio.
Figure 2.
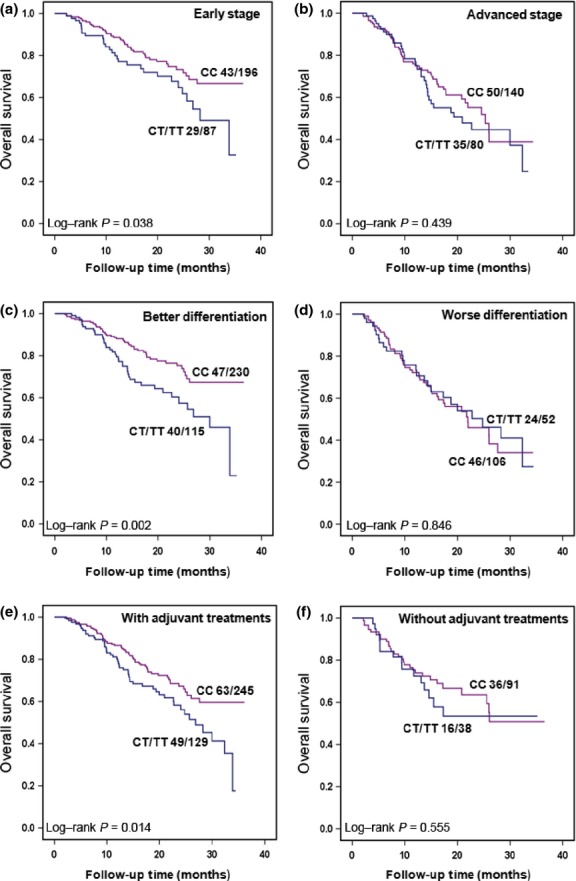
Kaplan–Meier plots of overall survival in Chinese patients with non-small-cell lung cancer with different genotypes of rs1126497 by stratified analysis in a dominant genetic model. The number of patients (deaths/total) is indicated for each subgroup. (a) Patients with stage I/II disease. (b) Patients with stage III/IV disease. (c) Patients with well or moderately differentiated tumors. (d) Patients with poorly differentiated or undifferentiated tumors. (e) Patients who received chemotherapy or radiotherapy. (f) Patients who did not receive chemotherapy or radiotherapy.
Discussion
In this study, we investigated the effects of two SNPs in the EpCAM gene on the survival of NSCLC patients. We found that a variant genotype (CT+TT) of rs1126497 in the EpCAM gene was significantly associated with poor prognosis (both OS and RFS) of NSCLC, whereas there was no significant association observed between the SNP rs1421 and the clinical outcomes of NSCLC patients. Further stratified analysis indicated that the effect of rs1126497 had more prominence in older patients, smokers, patients with squamous cell carcinoma, patients with well or moderately differentiated tumor, and patients who received adjuvant chemotherapy or radiotherapy treatment. Moreover, we examined the associations between these two SNPs and NSCLC progression by the logistic regression model and found that a variant genotype of rs1421 had significant association with advanced T-stage disease in NSCLC patients. To the best of our knowledge, this is the first study to investigate the association between EpCAM gene polymorphisms and NSCLC prognosis.
Previous studies have indicated that a functional polymorphism (rs1126497) in the EpCAM gene was associated with the risk of breast and cervical cancer.22,23 One of these studies showed that subjects carrying the CT+TT genotype had a 1.40-fold increased risk for breast cancer than patients with the CC genotype. Another study found that there was a dose–response of cancer risk for patients with the CT genotype (adjusted OR = 1.72) and TT genotype (adjusted OR = 1.96) for cervical cancer. Nonetheless, there is no report about the association between functional polymorphisms in the EpCAM gene and cancer prognosis. Our study found that variant genotypes of rs1126497 had a significant association with worse prognosis of NSCLC patients in both OS and RFS analyses, which is in concordance with those previous reports.
As a cell adhesion molecule, EpCAM is involved in cell adhesion and proliferation, which plays a regulative role in human cancer. It has been indicated that EpCAM is overexpressed in various types of cancers, including cancers of the colon and rectum,26 breast,27 prostate,28 bladder,11 and also frequently in lung cancer.12,29 Downregulation of EpCAM by RNAi decreases the proliferation, migration, and invasive capacity of breast cancer cells.30 In addition, EpCAM knockdown leads to substantial apoptosis in lung cancer cell lines rather than in a bronchial epithelial cell line.19 Moreover, recent studies have revealed that EpCAM plays a versatile role in carcinogenesis, in that it not only affects cell–cell adhesion but is also involved in many processes including cell migration, differentiation, metabolism.31 All these studies suggest that EpCAM play an essential role in the development of cancer, including lung cancer.
The EpCAM molecule consists of three domains: an extracellular domain with epidermal growth factor-like repeat and thyroglobulin repeat-like (TY) domain, a single transmembrane domain, and an intracellular domain (EpICD).32 The TY domain is capable of selectively discriminating cathepsin L and S, two similar cysteine proteases. It has the potential role of inhibiting the activity of cysteine proteases, and this activity depends on the specific interaction with target protein.33 The SNP rs1126497 is an ns polymorphism (M115T) located in the TY domain of EpCAM.32 Thus, the rs1126497 polymorphism could alter the primary structure of the TY domain, which may decrease the interaction between the TY domain and cathepsin L, weaken the inhibiting ability of cathepsin L, and thus eventually affect NSCLC patients' prognosis. More recently, many studies have focused on the EpCAM intracellular signaling pathway, in which EpCAM is cleaved by tumor necrosis factor-α converting enzyme (TACE/ADAM17) and γ-secretase and the ectodomain is released into the medium and EpICD into the cytoplasm.7,34 The released EpICD as a signal transducer is transported into the nucleus and forms a nuclear complex with FHL2, β-catenin, and Lef-1 to stimulate expression of target gene c-myc. This result supports the proliferative signaling activity of overexpressed EpCAM.34,35
In the stratified analysis, we observed that the significant associations between SNP rs1126497 and OS were only evident in NSCLC patients with better clinical parameters, such as early stage disease and well or moderately differentiated tumors, and in those receiving adjuvant chemotherapy or radiotherapy (Fig.2). To date, the detailed mechanism underlying the effect of SNP rs1126497 on cancer progression is largely unknown. However, this SNP has been reported to be significantly associated with risk of breast and cervical cancers in Chinese populations.22,23 It has also been reported that EpCAM is an early biomarker commonly overexpressed in many premalignant tissues, especially in epithelial neoplasia.36 These data suggest that this ns-SNP may play an important role in cancers at early stage. Moreover, it is generally accepted that most SNPs only have subtle effects on complex traits, such as cancers. This may partially explain why rs1126497 did not show a significant effect on prognosis in patients with worse clinical parameters, as the worse the cancer, the more complex and varied the disease. Additionally, a previous in vitro study suggested that EpCAM is a promising target to sensitize tumor cells to chemotherapy,37 which is in concordance with our finding that the effect conferred by this ns-SNP on prognosis was only evident in patients receiving adjuvant treatment. Taken together, it is reasonable to find that the effect of SNP rs1126497 on prognosis is more prominent in those patients with better clinical parameters and receiving adjuvant treatments.
Additionally, although our data did not show any association between rs1421 and NSCLC prognosis, we found that the rs1421 AG+GG genotypes were significantly associated with early T stage disease. This could be due to the location of rs1421 in miRNA binding sites of the EpCAM 3′-UTR region that may affect expression of EpCAM. Further study is needed to understand the effects of the rs1421polymorphism on tumor progression.
The biggest advantage of our study is that all subjects' characteristics were homogenous, and they were treated using similar procedures, which minimized the confounding effects. Nevertheless, there are limitations to our study. First, the follow-up time was relatively short. However, this limitation can be partially compensated by the high recurrence and death rate in our study population. Second, the relatively small sample size limited the analysis on the recessive model and caused an unstable estimation for the stratified analyses. A larger number of patients will be needed to assess the association between the SNP rs1126497 at EpCAM and NSCLC survival in future studies. Finally, functional assays are warranted to establish the underlying mechanism.
In summary, our study indicates for the first time that the SNP rs1126497 in the EpCAM gene is significantly associated with the prognosis of NSCLC. Our findings contribute to current understanding of the effects of EpCAM polymorphisms on NSCLC.
Acknowledgments
This work was supported by grant 81201583 from Program for New Century Excellent Talents in University of China and grant 2011ZX09307-001-04 from the National Key Scientific and Technological Project of China.
Disclosure Statement
The authors have no conflict of interest.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Sekido Y, Fong KM, Minna JD. Molecular genetics of lung cancer. Annu Rev Med. 2003;54:73–87. doi: 10.1146/annurev.med.54.101601.152202. [DOI] [PubMed] [Google Scholar]
- 4.Cho J. The International Association for the Study of Lung Cancer–the Lung Cancer Staging Project: better data, better decisions, better outcomes. Hawaii Med J. 2008;67:220–2. [PubMed] [Google Scholar]
- 5.Mulshine JL, Sullivan DC. Clinical practice. Lung cancer screening. N Engl J Med. 2005;352:2714–20. doi: 10.1056/NEJMcp042630. [DOI] [PubMed] [Google Scholar]
- 6.Chong JM, Speicher DW. Determination of disulfide bond assignments and N-glycosylation sites of the human gastrointestinal carcinoma antigen GA733-2 (CO17-1A, EGP, KS1-4, KSA, and Ep-CAM) J Biol Chem. 2001;276:5804–13. doi: 10.1074/jbc.M008839200. [DOI] [PubMed] [Google Scholar]
- 7.Munz M, Baeuerle PA, Gires O. The emerging role of EpCAM in cancer and stem cell signaling. Cancer Res. 2009;69:5627–9. doi: 10.1158/0008-5472.CAN-09-0654. [DOI] [PubMed] [Google Scholar]
- 8.van der Gun BT, Melchers LJ, Ruiters MH, de Leij LF, McLaughlin PM, Rots MG. EpCAM in carcinogenesis: the good, the bad or the ugly. Carcinogenesis. 2010;31:1913–21. doi: 10.1093/carcin/bgq187. [DOI] [PubMed] [Google Scholar]
- 9.Kimura H, Kato H, Faried A, et al. Prognostic significance of EpCAM expression in human esophageal cancer. Int J Oncol. 2007;30:171–9. [PubMed] [Google Scholar]
- 10.Went P, Dirnhofer S, Salvisberg T, et al. Expression of epithelial cell adhesion molecule (EpCam) in renal epithelial tumors. Am J Surg Pathol. 2005;29:83–8. doi: 10.1097/01.pas.0000.146028.70868.7a. [DOI] [PubMed] [Google Scholar]
- 11.Brunner A, Prelog M, Verdorfer I, Tzankov A, Mikuz G, Ensinger C. EpCAM is predominantly expressed in high grade and advanced stage urothelial carcinoma of the bladder. J Clin Pathol. 2008;61:307–10. doi: 10.1136/jcp.2007.049460. [DOI] [PubMed] [Google Scholar]
- 12.Pak MG, Shin DH, Lee CH, Lee MK. Significance of EpCAM and TROP2 expression in non-small cell lung cancer. World J Surg Oncol. 2012;10:53. doi: 10.1186/1477-7819-10-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Went P, Vasei M, Bubendorf L, et al. Frequent high-level expression of the immunotherapeutic target Ep-CAM in colon, stomach, prostate and lung cancers. Br J Cancer. 2006;94:128–35. doi: 10.1038/sj.bjc.6602924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ni J, Cozzi PJ, Duan W, et al. Role of the EpCAM (CD326) in prostate cancer metastasis and progression. Cancer Metastasis Rev. 2012;31:779–91. doi: 10.1007/s10555-012-9389-1. [DOI] [PubMed] [Google Scholar]
- 15.Munz M, Kieu C, Mack B, Schmitt B, Zeidler R, Gires O. The carcinoma-associated antigen EpCAM upregulates c-myc and induces cell proliferation. Oncogene. 2004;23:5748–58. doi: 10.1038/sj.onc.1207610. [DOI] [PubMed] [Google Scholar]
- 16.Seligson DB, Pantuck AJ, Liu X, et al. Epithelial cell adhesion molecule (KSA) expression: pathobiology and its role as an independent predictor of survival in renal cell carcinoma. Clin Cancer Res. 2004;10:2659–69. doi: 10.1158/1078-0432.ccr-1132-03. [DOI] [PubMed] [Google Scholar]
- 17.Songun I, Litvinov SV, van de Velde CJ, Pals ST, Hermans J, van Krieken JH. Loss of Ep-CAM (CO17-1A) expression predicts survival in patients with gastric cancer. Br J Cancer. 2005;92:1767–72. doi: 10.1038/sj.bjc.6602519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ensinger C, Kremser R, Prommegger R, Spizzo G, Schmid KW. EpCAM overexpression in thyroid carcinomas: a histopathological study of 121 cases. J Immunother. 2006;29:569–73. doi: 10.1097/01.cji.0000211303.52503.da. [DOI] [PubMed] [Google Scholar]
- 19.Hase T, Sato M, Yoshida K, et al. Pivotal role of epithelial cell adhesion molecule in the survival of lung cancer cells. Cancer Sci. 2011;102:1493–500. doi: 10.1111/j.1349-7006.2011.01973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehne B, Lewis CM, Schlitt T. From SNPs to genes: disease association at the gene level. PLoS ONE. 2011;6:e20133. doi: 10.1371/journal.pone.0020133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Y, Spitz MR, Amos CI, Lin J, Schabath MB, Wu X. An evolutionary perspective on single-nucleotide polymorphism screening in molecular cancer epidemiology. Cancer Res. 2004;64:2251–7. doi: 10.1158/0008-5472.can-03-2800. [DOI] [PubMed] [Google Scholar]
- 22.Hu M, Jian L, Zhang L, et al. Functional polymorphism in the EpCAM gene is associated with occurrence and advanced disease status of cervical cancer in Chinese population. Mol Biol Rep. 2012;39:7303–9. doi: 10.1007/s11033-012-1560-9. [DOI] [PubMed] [Google Scholar]
- 23.Jiang L, Zhang C, Li Y, et al. A non-synonymous polymorphism Thr115Met in the EpCAM gene is associated with an increased risk of breast cancer in Chinese population. Breast Cancer Res Treat. 2011;126:487–95. doi: 10.1007/s10549-010-1094-6. [DOI] [PubMed] [Google Scholar]
- 24.Tomita N, Yamano T, Matsubara N, Tamura K. A novel genetic disorder of Lynch syndrome – EPCAM gene deletion. Gan To Kagaku Ryoho. 2013;40:143–7. [PubMed] [Google Scholar]
- 25.Sivagnanam M, Mueller JL, Szigeti R, Gopalakrishna GS, Kellermayer R. Transcriptional read-through induction treatment trial in intestinal failure induced by an EpCAM nonsense mutation. Case Report Med. 2012;2012:173195. doi: 10.1155/2012/173195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herlyn M, Steplewski Z, Herlyn D, Koprowski H. Colorectal carcinoma-specific antigen: detection by means of monoclonal antibodies. Proc Natl Acad Sci USA. 1979;76:1438–42. doi: 10.1073/pnas.76.3.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt M, Hasenclever D, Schaeffer M, et al. Prognostic effect of epithelial cell adhesion molecule overexpression in untreated node-negative breast cancer. Clin Cancer Res. 2008;14:5849–55. doi: 10.1158/1078-0432.CCR-08-0669. [DOI] [PubMed] [Google Scholar]
- 28.Poczatek RB, Myers RB, Manne U, et al. Ep-Cam levels in prostatic adenocarcinoma and prostatic intraepithelial neoplasia. J Urol. 1999;162:1462–6. [PubMed] [Google Scholar]
- 29.Piyathilake CJ, Frost AR, Weiss H, Manne U, Heimburger DC, Grizzle WE. The expression of Ep-CAM (17-1A) in squamous cell cancers of the lung. Hum Pathol. 2000;31:482–7. doi: 10.1053/hp.2000.6711. [DOI] [PubMed] [Google Scholar]
- 30.Osta WA, Chen Y, Mikhitarian K, et al. EpCAM is overexpressed in breast cancer and is a potential target for breast cancer gene therapy. Cancer Res. 2004;64:5818–24. doi: 10.1158/0008-5472.CAN-04-0754. [DOI] [PubMed] [Google Scholar]
- 31.Munz M, Zeidler R, Gires O. The tumour-associated antigen EpCAM upregulates the fatty acid binding protein E-FABP. Cancer Lett. 2005;225:151–7. doi: 10.1016/j.canlet.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 32.Baeuerle PA, Gires O. EpCAM (CD326) finding its role in cancer. Br J Cancer. 2007;96:417–23. doi: 10.1038/sj.bjc.6603494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guncar G, Pungercic G, Klemencic I, Turk V, Turk D. Crystal structure of MHC class II-associated p41 Ii fragment bound to cathepsin L reveals the structural basis for differentiation between cathepsins L and S. EMBO J. 1999;18:793–803. doi: 10.1093/emboj/18.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maetzel D, Denzel S, Mack B, et al. Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell Biol. 2009;11:162–71. doi: 10.1038/ncb1824. [DOI] [PubMed] [Google Scholar]
- 35.Carpenter G, Red Brewer M. EpCAM: another surface-to-nucleus missile. Cancer Cell. 2009;15:165–6. doi: 10.1016/j.ccr.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 36.Patriarca C, Macchi RM, Marschner AK, Mellstedt H. Epithelial cell adhesion molecule expression (CD326) in cancer: a short review. Cancer Treat Rev. 2011;38:68–75. doi: 10.1016/j.ctrv.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Hussain S, Pluckthun A, Allen TM, Zangemeister-Wittke U. Chemosensitization of carcinoma cells using epithelial cell adhesion molecule-targeted liposomal antisense against bcl-2/bcl-xL. Mol Cancer Ther. 2006;5:3170–80. doi: 10.1158/1535-7163.MCT-06-0412. [DOI] [PubMed] [Google Scholar]


