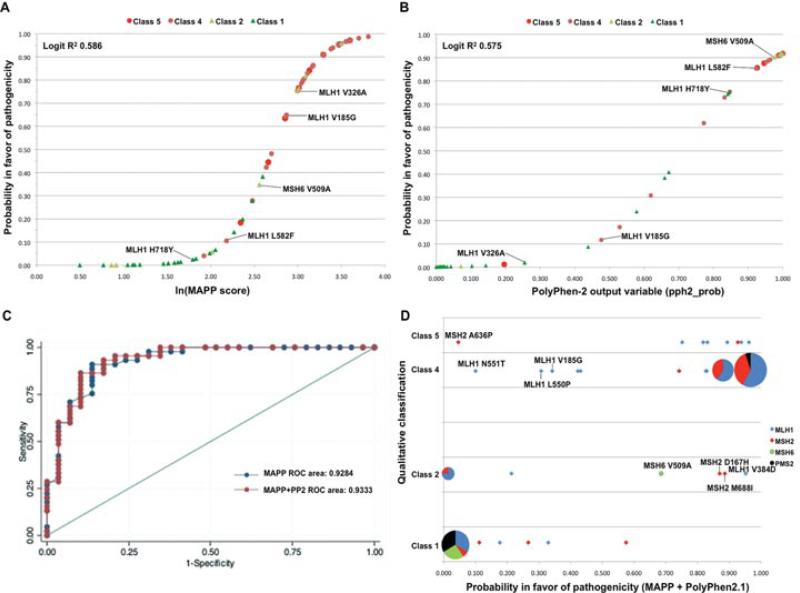
Figure 1.
A: Distribution of the set of 74 class 1–2–4–5 missense variants on a sigmoid curve for MAPP outputs derived using the logit regression calibration equation—Logit(Pr) = −10.84 + 3.99(ln[MAPP score]), where Pr is the probability in favor of pathogenicity. The classifications of the variants are identified by colored symbols (defined in the legend). B: Sigmoid curve showing the outputs of the 74 missense variants from PolyPhen-2.1 using the logit regression calibration equation—Logit(Pr) = −6.04 + 8.45(pph2_prob). The missense variants with the largest differences (Δ) in probability of pathogenicity derived from MAPP versus PolyPhen-2.1 are identified in (A) and (B): MLH1 V185G (Δ = 0.528), MLH1 V326A (Δ = 0.733), MLH1 L582F (Δ = 0.748), MLH1 H718Y (Δ = 0.719), and MSH6 V509A (Δ = 0.553). C: Receiver operating characteristic (ROC) curves for MAPP and combined MAPP + PolyPhen-2.1 accuracy, using probabilities and scores associated with each program. D: Scatter diagram comparing the qualitatively assigned probability in favor of pathogenicity (class 1—not pathogenic, class 2—likely not pathogenic, class 4—likely pathogenic, and class 5—pathogenic) with the probability of pathogenicity as a continuous variable calculated from the MAPP + PolyPhen-2.1 outputs. The pie graphs are proportional representations of overlapping variants in the four mismatch repair genes (MLH1—blue, MSH2—red, MSH6—green, and PMS2—black). The variants with in silico scores predicting <0.40 probability of pathogenicity that were qualitatively classified as class 4 and class 5 are identified, as were variants that had in silico scores predicting >0.60 probability of pathogenicity with qualitative classifications of class 1 and class 2.