Abstract
Shape complementarity is an important component of molecular recognition, and the ability to precisely adjust the shape of a binding scaffold to match a target of interest would greatly facilitate the creation of high affinity protein reagents and therapeutics. Here we describe a general approach to control the shape of the binding surface on repeat protein scaffolds, and apply it to leucine rich repeat proteins. First, a set of self-compatible building block modules are designed that when polymerized each generate surfaces with unique but constant curvatures. Second, a set of junction modules that connect the different building blocks are designed. Finally, new proteins with custom designed shapes are generated by appropriately combining building block and junction modules. Crystal structures of the designs illustrate the power of the approach in controlling repeat protein curvature.
Repeat protein scaffolds have attracted much attention as alternative binding scaffolds to antibodies1-4 and also as building blocks of protein nanomaterials5-7 because of their intrinsic modularity and high stability. The leucine rich repeat (LRR) is a repeat protein scaffold with a horseshoe-like global structure in which the concave surface is often a binding interface8. While LRRs share a common structural motif (LxxLxLxxN/C), different LRR modules generate proteins with distinct global curvatures when the repeat modules are packed on themselves9. Irregular LRR modules are frequently observed interspersed within arrays of canonical repeat modules; their presence contributes to the curvature diversity within the family. For example, Toll-like receptor 4 (TLR4) contains three distinct regions of LRR repeats, each having different curvatures which collectively generate a surface with high shape complementarity to the target surface of the MD2 protein10. Current engineering approaches have focused on changing residues at the binding surfaces of an already existing or consensus repeat protein11-16, varying the numbers of repeat modules17-19, and fusing naturally occurring repeat proteins10,20,21. While powerful, these strategies do not allow customization of repeat protein curvature for a specific application.
Here, we describe a general computational design strategy to create new repeat proteins with custom-specified curvature. We demonstrate the power of the approach by designing twelve novel proteins with different curvatures. Crystal structures show that the method allows control of repeat protein curvature with atomic-level accuracy.
Results
Strategy for curvature-tunable scaffold design
Our design strategy has three steps (Fig. 1a). The first step is the design of a set of idealized self-compatible building block modules (BB1, BB2, …., BBn) from which a series of proteins of variable length BBin can be created directly by varying the number of building block repeats without any further engineering. These “homo-building block” proteins will have a constant curvature defined by the base building block module. The second step is the design of a set of junction modules (JNBBi→BBj) that connect building block module i to building block module j. A critical feature of the design at step one and two is that the interfaces between individual building blocks, as well as those between building blocks in junction modules, have sufficiently low energy that the orientation between all units depends only upon the identity of adjacent repeats and is independent of the longer-range context. This enables the third and final step -- general module assembly -- the combination of building blocks and junction modules to generate a protein with a desired overall curvature. While the overall strategy is applicable to any repeat protein, in this paper we focus on LRRs. We describe the computational design and experimental characterization for each step in the following sections.
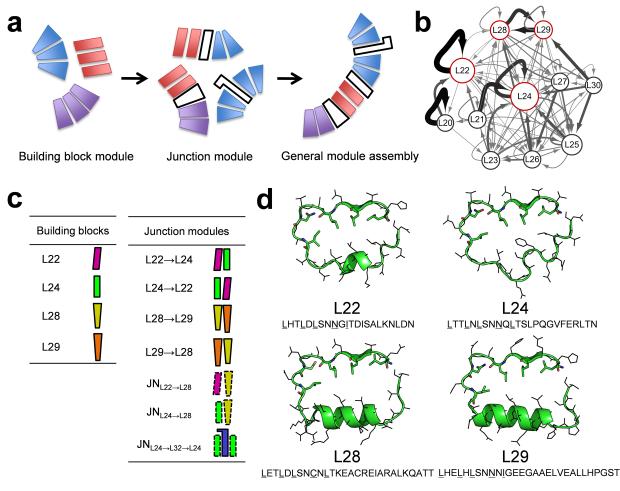
Figure 1.
(a) Overview of curvature-tunable scaffold design: idealized building block module design, junction module design, and general module assembly. (b) Module organization of natural LRR modules is represented by a network where nodes represent modules and edges transitions between modules. The size of nodes and the thickness of edges are proportional to the frequencies observed in the PDB. (c) Graphical representation of building block and junction modules. (d) Idealized building block module structures and sequences. The highly conserved residues are shown in sticks.
Step 1: Building block module selection and design
Nature provides a diverse set of LRR modules, with lengths from 20 to 30 amino acids8, but only a few possess high self-compatibility such that repeated stacking of the same module generates a well-folded protein structure. We generated a Markov transition model for naturally occurring LRR proteins to investigate the overall patterns of module organization in LRR structures. In the model, nodes correspond to individual modules (represented by the module length: L22 indicates an LRR module with 22 residues, etc) and edges to transitions between modules with strength proportional to the transition frequency observed between the modules in the Protein Data Bank (PDB) (see Methods section). The resulting transition network (Fig. 1b) has strong self-edges corresponding to packing of identical modules for L22 and L24, and strong mutual transitions between L28 and L29. Accordingly, we selected these LRR types to design the idealized building blocks (Fig. 1c-d).
A recently developed Rosetta repeat protein idealization method22 was used to design ideal versions of each unit. Different instances of the naturally occurring repeat units have somewhat variable sequences; the idealization process generates a single low energy repeat unit (both sequence and structure) guided by the available information for the family. Briefly, an idealized poly-valine backbone structure with identical repeats was generated using RosettaRemodel23 with LRR family-specific constraints. Rosetta sequence design guided by a family-specific sequence profile was then carried out, while constraining the sequences to be identical for each repeat. The idealization of the L24 module (DLRR_B) is described in Parmeggiani et al.22 We applied the idealization procedure to the L22 module (DLRR_A) and the two-unit {L28→L29} module (DLRR_C), and obtained the sequences and models in Figure 1d.
Genes were synthesized for proteins containing 5 to 7 idealized building block modules. The N–terminal capping domain of internalin B was fused to DLRR_A and DLRR_B to enhance protein solubility and expression12,20 whereas DLRR_C was expressed without a capping motif; instead the sequences of the N and C terminal repeats were redesigned to eliminate exposed hydrophobic residues. The idealized repeat designs were expressed in E.coli and found to be soluble and to have high thermal stability (Fig. 2c).
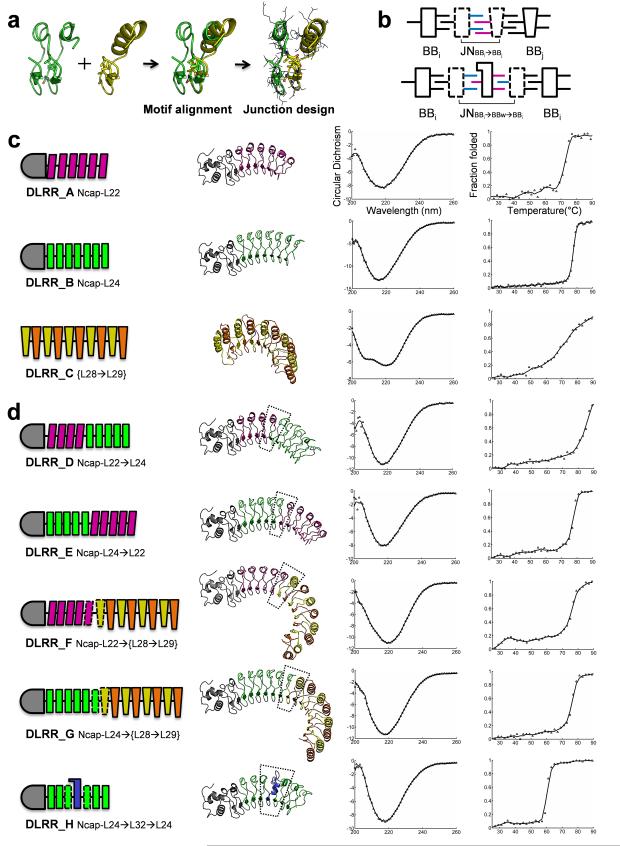
Figure 2.
Building block and junction module design. (a) Junction module design illustrated by the L24→L28 fusion design. (b) Schematic representing a two-unit junction module JNBBi→BBj and a three-unit wedge module JNBBi→BBw→BBi. (c) The idealized building block modules for L22, L24, and {L28→L29}. (d) The junction modules for L22→L24, L24→L22, L22→L28 (DLRR_F6), L24→L28 (DLRR_G2), and L24→L32→L24 (DLRR_H2). For each design, module organization, model structure, far-UV CD spectra (mean residue ellipticity, [θ] × 10−3 deg cm2 dmol−1), and thermal denaturation profile following the wavelength 218 nm are displayed from left to right. When multiple designs exist, data of the most stable one are shown. The backbone modules are indicated in different colors: magenta-L22, green-L24, yellow-L28, and orange-L29 (Fig. 1c). The designed junction modules are represented by dotted lines, and the N-terminal capping domain connected to L22 or L24 is shown in gray.
We solved the crystal structures of DLRR_A (L226) and DLRR_B (L247) (Table 1) and found that they closely match the design models (DLRR_A at Cα root mean square deviation (RMSD) 1.4 Å; DLRR_B at Cα RMSD 1.7 Å, Fig. 3a-b). The crystal structures contain water-mediated networks localized to the convex side of the repeats; it may be possible to incorporate these in future design calculations (Supplementary Fig. 1a). Each of the idealized building block repeats has the expected overall curvature: repeats of the L22 and L24 building blocks generate solenoid-like structures, whereas repeats of the {L28→L29} building block are almost circular and have a more curved concave surface. Parametric descriptions of the global shapes generated by each building block repeat are provided in Supplementary Figure 1b and Supplementary Table 1.
Table 1.
Data collection and refinement statistics (molecular replacement)
| Crystal | DLRR_A | DLRR_E | DLRR_G3 | DLRR_H2 | DLRR_I | DLRR_K |
|---|---|---|---|---|---|---|
| Data collection | ||||||
| Space group | P21 | P212121 | F222 | P212121 | C2 | P22121 |
| Cell dimensions | ||||||
| a, b, c (Å) | 57.66, 245.07, 57.73 | 32.12,77.71,101.89 | 91.13, 136.38, 161.74 | 89.78,96.50,136.36 | 109.49, 42.71, 67.82 | 36.87, 93.37, 126.24 |
| α,β,υ(°) | 90, 115.36, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 102.4, 90 | 90, 90, 90 |
| Resolution (Å) | 50(2.36)* | 42.6(1.93) | 23.5(2.53) | 50(2.9) | 50(1.73) | 50(2.8) |
| R sym | 0.081(0.183) | 0.063(0.171) | 0.067(0.153) | 0.092(0.529) | 0.076(0.252) | 0.192(0.742) |
| I/σI | 24.0(6.4) | 17.7(8.0) | 15.5(4.1) | 17.2(3.85) | 33.7(4.5) | 8.9(2.3) |
| Completeness (%) | 96.7(83.7) | 99.8(96.0) | 98.1(85.5) | 99.8(99.6) | 96.3(83.7) | 99.7(99.2) |
| Redundancy | 5.7(3.0) | 6.4(5.0) | 4.5(1.9) | 7.2(7.1) | 10.3(2.3) | 6.2(5.8) |
| Refinement | ||||||
| Resolution (Å) | 50(2.36) | 42.6(1.93) | 23.5(2.53) | 50(2.9) | 50(1.73) | 50(2.8) |
| No. reflections | 34180 | 19993 | 17061 | 25484 | 31150 | 10729 |
| Rwork(%) | 18.9(22.3) | 15.86(17.50) | 18.47(23.4) | 21. 16(32.8) | 17.07(21.50) | 20.75(28.4) |
| Rfree (%) | 24.2(27.7) | 22.38(23.7) | 24.65(36.1) | 25.15(48.5) | 21.99(31.70) | 28.53(36.0) |
| No. atoms | ||||||
| Protein | 6771 | 2388 | 3456 | 7841 | 2577 | 3582 |
| Ligand/ion | 8 | 12 | 29 | 20 | --- | 1 |
| Water | 230 | 106 | 96 | 1 | 199 | 18 |
| B-factors | ||||||
| Protein | 12.13 | 14.65 | 11.73 | 69.14 | 10.84 | 16.96 |
| Ligand/ion | 35.53 | 39.67 | 54.64 | 85.26 | --- | 42.89 |
| Water | 18.0 | 35.39 | 21.91 | 50.44 | 25.43 | 18.60 |
| r.m.s. deviations | ||||||
| Bond length (Å) | 0.0137 | 0.0181 | 0.0138 | 0.0126 | 0.0194 | 0.0136 |
| Bond angles (°) | 1.661 | 1.810 | 1.629 | 1.651 | 2.052 | 1.475 |
Values in parentheses are for highest-resolution shell.
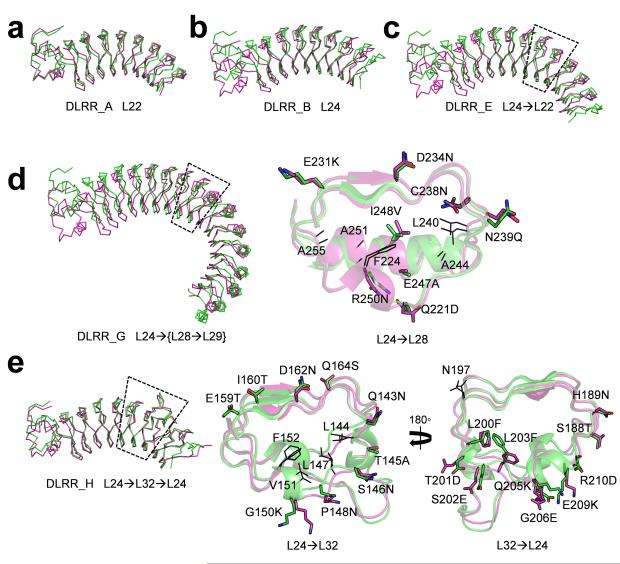
Figure 3.
Crystal structures of the idealized building block and junction module designs. The design models (green) for (a) L22, (b) L24, (c) L24→L22, (d) L24→L28 (DLRR_G3), and (e) L24→L32→L24 (DLRR_H2) superimposed on the crystal structures (magenta). Close-up views for the designed junction modules (dashed region) are shown for (d) and (e). Residues mutated from the original building block sequences are annotated and shown as sticks. Additional residues that vary within the designs are shown as lines. Except for that of DLRR_A, the crystal structures have missing electron density at the C-terminus (10–20 amino acids). PyMOL (http://www.pymol.org/) are used in all structural visualization.
Step2: Design of junction modules
We devised a computational protocol for junction module design which takes advantage of the conserved motif (LxxLxLxxN/C) in the idealized LRR building blocks: the core residues are kept constant to maintain a stable hydrophobic core, while the evolutionarily variable positions, primarily located on the convex side, are optimized to create a low energy interface between adjacent modules. To generate a junction module JNBBi→BBj connecting building block i and building block j, we start from a two-unit BBi2 module and a one-unit BBj module (Fig. 2a). The second unit in BBi2 is superimposed on BBj by aligning the core motif residues. RosettaCM24 is then used to generate a hybrid structure BBi→BBj with coordinates based on those of the first unit in BBi2 before the core motif and those of BBj after the motif. The residues at the fusion interface are optimized using RosettaDesign25. This redesigned hybrid two-unit structure BBi→BBj is the junction module JNBBi→BBj between building block i and building block j (Fig. 2b).
A special case of a junction module is a three-unit module JNBBi→BBw→BBi that connects two identical copies of the same building block but has a structure different than that of the building block (Fig. 2b). We call such junction modules between two identical building blocks ‘wedge’ modules. Like other junction modules, wedge modules produce a local change in the protein curvature. We designed and characterized five junction modules connecting the building block modules described in the previous section.
L22→L24 and L24→L22. A junction module for L22→L24 was generated previously without hydrophobic core design12, and hence direct fusion constructs between L22 and L24 were made in both directions (i.e. L22→L24 and L24→L22) to test compatibility between the two idealized modules. The hybrid model structures showed high structural compatibility without further design. Hence the junction modules in these cases are simply the fusion of the two building blocks.
Two fusion proteins for L22→L24 (DLRR_D) and L24→L22 (DLRR_E) were expressed in E. coli, and found to be soluble and monomeric in Size-Exclusion Chromatography coupled to Multiple Angle Light Scattering (SEC–MALS) experiments (Supplementary Fig. 1e). Far-UV Circular Dichroism (CD) spectra and thermal denaturation profiles suggested well-packed structures with the expected secondary structure content (Fig. 2d). The fusion proteins had similar or higher stability than the original L22 (DLRR_A) or L24 (DLRR_B) designs (Table 2); L22 and L24 evidently have high compatibility despite the rare occurrence of fusions between them in nature (Fig. 1b). The crystal structure for L24→L22 (DLRR_E) was determined at 1.9 Å resolution and showed high consistency with the design model (Fig. 3c) and the original L22 or L24 structures (Supplementary Fig. 1c).
Table 2.
Summary of fusion designs and experimental results
| Design Name | Module organization§ | Modules¶
(repeat units) |
designs tested |
Soluble | Folded (CD) |
Mono meric |
X- ray |
Tm
(°C) |
RMSD1 | RMSD2 |
|---|---|---|---|---|---|---|---|---|---|---|
| DLRR_A | Ncap–L226 | 6 (6) | 1 | 1 | 1 | 1 | 1 | 73 | 1.4 (0.8, 2.0) |
0.4 (0.5, 1.0) |
| DLRR_BΔ | Ncap–L247 | 7 (7) | 1 | 1 | 1 | 1 | 1 | 78 | 1.7 (1.5, 2.9) |
0.3 (0.6, 0.4) |
| DLRR_C | {L28→L29}5 | 5 (10) | 5 | 5 | 1 | # | 71 | |||
| DLRR_D | Ncap–L224→L245 | 9 (9) | 1 | 1 | 1 | 1 | 87 | |||
| DLRR_E | Ncap-L245→L225 | 10 (10) | 1 | 1 | 1 | 1 | 1 | 77 | 2.1 (1.4, 2.0) |
0.4 (0.7, 0.7) |
| DLRR_F | Ncap–L224–JNL22→L28→L29→{L28→L29}3 | 9 (13) | 6 | 6 | 6 | 6 | 77 | |||
| DLRR_G | Ncap–L245–JNL24→L28→L29→{L28→L29}3 | 10 (14) | 6 | 6 | 6 | 6 | 1 | 81 | 2.6 (3.1, 3.8) |
0.8 (0.8, 2.2) |
| DLRR_H | Ncap–L242–JNL24→L32→L24–L242 | 5 (7) | 4 | 4 | 4 | 2 | 1 | 65 | 0.9 (0.5, 1.0) |
0.8 (0.7, 1.2) |
| DLRR_I | Ncap–L242–JNL24→L32→L24–JNL24→L32→L24–L242 | 6 (10) | 1 | 1 | 1 | 1 | 1 | 53 | 1.7 (1.2, 2.3) |
0.5 (0.5, 0.7) |
| DLRR_J | Ncap–L224→L242–JNL24→L28→L29→{L28→L29}2 | 10 (13) | 1 | 1 | 1 | 1 | 82 | |||
| DLRR_K | Ncap–L242–JNL24→L32→L24–L243– JNL24→L28→L29→{L28→L29}2 |
10 (15) | 1 | 1 | 1 | 1 | 1 | 75 | † | 1.1 (1.2, 3.9) |
| DLRR_L | Ncap–L223→L243–JNL24→L32→L24–L243
JNL24→L28→L29→{L28→L29}2 |
14 (19) | 1 | 1 | 1 | 1 | 83 |
The superscripts represent the number of repeat units
Alternatively occurring two-unit {L28→L29} is considered as one module.
Experimental data of DLRR_B is from Parmeggiani et al.22
DLRR_C forms a dimer
Tm is estimated by calculating the infection point of the melting curve at 218 nm, and the highest Tm value is represented when multiple designs exist.
RMSD1 : Cα RMSD (Å) between crystal structure and model generated from design models of building blocks and junction modules.
RMSD2 : Cα RMSD (Å) between crystal structure and model generated using crystal structures of building block and junction modules (Supplementary Fig. 5c)
RMSD values for the first and the last unit in global structure alignment are provided in parenthesis.
Model of DLRR_K is generated by module assembly without an initial design model.
L22→L28 and L24→L28. Designs of junction modules for L22→L28 and L24→L28 are challenging because of substantial differences in their structures, including their module length (22 or 24 vs. 28), their secondary structure in the variable region (310-helix or loop vs. α-helix), their curvature on the concave surface (moderately-curved vs. highly-curved) and their global shape (super-helical vs. circular). The initial fusion models generated by RosettaCM24 (before redesign) contained side chain clashes and cavities at the interface between the modules (Supplementary Fig. 1d). Residues at the fusion interface were therefore redesigned to improve the all-atom Rosetta energy and packing as assessed by RosettaHoles26. The junction designs were based solely on building block models generated by Rosetta25 because the crystal structures of the building blocks were not determined. Six designs for L22→L28 (DLRR_F) and six designs for L24→L28 (DLRR_G) were experimentally characterized (Table 2).
All designs were expressed in E. coli and found to be highly soluble and monomeric in SEC–MALS experiments (Supplementary Fig. 2 and 3). They displayed well-defined far-UV CD spectra with minimum near 218 nm, similar to those of previously characterized LRRs with primarily beta sheet secondary structure. Thermal denaturation experiments showed cooperative unfolding for all fusion designs (Fig. 2d), suggesting a well-packed hydrophobic core. Fusion of more stable LRR modules to less stable LRR modules via a well-designed junction appears to increase overall stability: the stability of all the junction module containing designs was greater than that of the original {L28→L29}5 design (DLRR_C).
We determined the crystal structure of the L24→L28 fusion (DLRR_G3) to evaluate the accuracy of the design. The crystal structure, determined at 2.5 Å resolution, shows the atomic details of the junction module as well as the structures of L24 and {L28→L29} modules (Fig. 3d). The assumption underlying our approach that curvature can be locally controlled is supported by the similarity of the L24 modules (Cα RMSD 0.3 Å) in DLRR_G3 structure to those in the all L24 DLRR_B structure, and by the similarity of the {L28→L29} modules (Cα RMSD 1.3 Å) to the {L28→L29} modules in DLRR_C model. The key core side chain interactions in the junction module are very similar in the design model and crystal structure (Cα RMSD 0.9 Å, Fig. 3d).
L24→L32→L24. In addition to the junction modules linking the different building block modules, we designed a wedge module inserted between L24 modules. In native LRR proteins, inserting an ‘irregular’ module between the regular modules is a common way to generate structural diversity by altering the overall curvature or forming a binding interface other than the concave surface (for example, the diverse LRR module organization and irregular binding surfaces in TLR family27 and plant LRR proteins28). The idealized L24 repeat structure (DLRR_B) was chosen as a base scaffold because it had the highest stability among the three idealized LRRs.
For the wedge module design, L24→[any-length-of-LRR]→L24 triples were retrieved from the LRRML database29 to identify irregular modules flanked by the L24 modules. A total of 21 unique irregular modules were identified. We selected the 32-length LRR unit (L32) found in the Toll-like receptor 3 structure30 (PDB ID: 2A0Z, 532–563) as a starting point. L32 has a relatively rigid and structured loop located on its convex surface which could be useful in future binding pocket designs. The junction module design process was applied to the two fusion interfaces (L24→L32 and L32→L24) which resulted in the wedge module JNL24→L32→L24 (DLRR_H). Four designs for L24→L32→L24 were selected and experimentally characterized (Table 2).
All designs were expressed in E.coli and found to be soluble. Two designs were monomeric in SEC-MALS experiment. Thermal denaturation experiments showed that insertion of the wedge module generally decreased stability of the base scaffold, but unfolding was still cooperative (Fig. 2d; Supplementary Fig. 4). The crystal structure of DLRR_H2 determined at 2.9 Å resolution was consistent to the design model (Cα RMSD 0.9 Å), confirming the accuracy of the junction module design protocol (Fig. 3e).
Step 3: Curvature specification by general module assembly
The crystal structures described thus far demonstrate that the building block modules (L22, L24, L28, and L29), junction modules (L22→L24, L24→L22, L24→L28, L28→L29, and L29→L28) and wedge modules (L24→L32→L24) all have structures that are very similar to the design models regardless of overall protein context. In principle, this enables design of combinations of modules to achieve a desired curvature. We represent the space of possible LRR structures as a network consisting of building block modules (nodes) connected by junction modules (edges) as in Figure 1b (Fig. 4a). Any sequence of modules generated by following the edges in the network corresponds to an LRR structure with unique curvature. For example, all the 18,786 possible fusion structures consisting of 12 modules are depicted in Figure 4b as lines connecting the center of masses for each repeat module in the structure. The curvature diversity is orders of magnitude greater than that of the original LRRs containing the same number of building block modules.
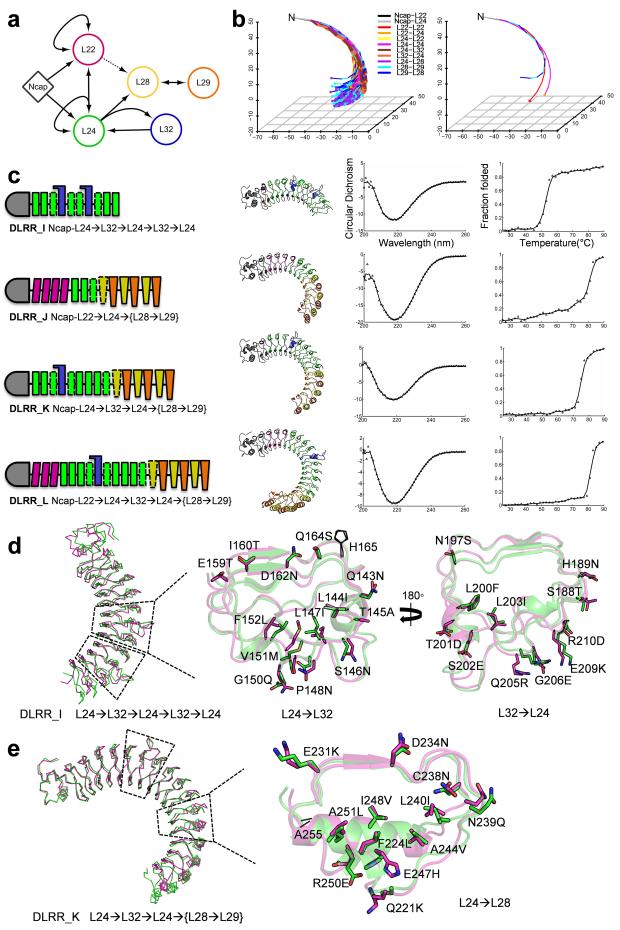
Figure 4.
Control of curvature by general module assembly (a) Network representing the generation of a new LRR structure using the idealized building block modules (nodes) and the designed junction modules (edges). The edge for L22→L28 is represented by a dotted line due to the lack of crystal structure, and not used in the module assembly. (b) Curvature diversity of general 12-unit LRR structures (left) is compared to that of the three building block structures (right). The LRR structures are generated by arbitrary module assembly based on the network (a), and the lines connecting the center of masses for each module are only shown for clarity. The structures are aligned based on the N-cap domain, and the C-termini are represented by dots colored by the relevant LRR modules. (c) Module organization, model structure, far-UV CD spectra, and thermal denaturation results for the four multiple fusion designs. (d,e) Comparison between the design model (green) and the crystal structure (magenta) for DLRR_I (d) and DLRR_K (e) with a close-up view of the fusion interfaces. Since the two L32 modules in DLRR_I are nearly identical with a Cα RMSD 0.2 Å, only the first L32 is shown.
We chose to use models of the individual building block and junction modules extracted from the crystal structures described thus far in the general module assembly process rather than the original design models of these units. While the building block modules are similar to previously described structures, the designed junction modules have sequences (Supplementary Fig. 5a) and structures (Supplementary Fig. 5b) quite different from previously described LRRs. Because of the imperfect state of computational protein design, we consider the crystal structures (which differ from the design models by Cα RMSD 0.2–1.0 Å) to be more accurate representations of the structures these modules are likely to adopt in new designs (Supplementary Fig. 5c).
General module assembly and experimental characterization
As a proof-of-concept for general module assembly, we designed four multiple fusion constructs (DLRR_I, DLRR_J, DLRR_K, and DLRR_L in Fig. 4c). The designs contain more than two fusion interfaces which result in large super-helical structures comparable in size to TLR431 (PDB ID: 3FXI, 626 residues) and plant steroid receptor BRI132 (PDB ID: 3RIZ, 743 residues). The module organization and module origins for each design are shown in Table 2, Figure 4c, and Supplementary Table 2.
Experimental characterization showed that the general module assembly protocol is quite robust. All of the multiple fusion designs were expressed in E. coli and found to be soluble, monomeric with well-defined CD spectra, cooperative unfolding transitions, and high thermal stability (Fig. 4c; Supplementary Fig. 1e). This is notable as all are quite large and complex proteins. We succeeded in solving the crystal structures of two of the designs as described in the following two paragraphs.
Design DLRR_I contains two successive L32 wedge modules with multiple flanking N and C-terminal L24 modules. We solved the crystal structure of DLRR_I at 1.7 Å resolution (Fig. 4d). Consistent with the assumption of context independent structure of the individual modules, the two L32 wedge modules in DLRR_I and the single L32 wedge module in DLRR_H2 are nearly identical over the backbone and core side chains (Cα RMSD 0.3–0.5 Å). Over the full 10 repeat unit structure, the crystal structure is closer to the model (Cα RMSD 0.5 Å, Supplementary Fig. 5c) assembled from the crystal structures of the individual building block and junction modules extracted from DLRR_B and DLRR_H than to the model (Cα RMSD 1.7 Å, Fig. 4d) assembled from the design models of the individual modules (Table 2), supporting our decision to use the crystal structures of the building blocks rather than the original design models in the general module assembly calculations.
Design DLRR_K consists of two L24 modules followed by the L32 module, three additional L24 modules, the L24→L28 junction module, and three {L28→L29} modules--a total of 15 repeat units. Such complexity of module organization is rarely if ever observed in naturally occurring LRRs. The protein is monomeric and stable, with a Tm of 75 °C. The crystal structure of DLRR_K at 2.8 Å resolution is very close to the general module assembly model (built from crystal structures of the individual modules from previous structures), with a Cα RMSD of 1.1 Å (Fig. 4e).
Taken together, these data suggest that general module assembly based on designed building block and junction modules can produce new structures with predefined shapes with high robustness and accuracy.
Discussion
We have described a general approach to creating repeat proteins with custom-designed shape through combination of designed building block and junction modules. The generation of scaffolds with defined curvatures using our computational approach is very likely simpler than that which occurred during the complex evolution of naturally occurring LRRs, and is considerably more controlled than what can be achieved in library selection approaches. The strategy allows the ready programming of a rich diversity of scaffolds with distinct curvatures: over 18,000 distinct 12 repeat unit structures can in principle be generated with our current set of building block and junction modules (Fig. 4b; Supplementary Table 3). The stable and well-expressed DLRR_L design (Fig. 4c) has a complex organization with five different types of modules (19 repeat units) in total; for this length there are over 5,000,000 distinct possibilities with our current module set, and increasing the repertoire of idealized building block and junction module designs would enrich the curvature diversity still further.
Our approach integrates protein structural analysis with energy driven design calculations to arrive at the idealized building block and junction modules, and computation and experiment to achieve high accuracy models of the complex repeat proteins generated by the module assembly process. While a completely energy driven approach would be preferable on aesthetic grounds, making use of information extracted from naturally occurring LRRs and from the crystal structures of idealized LRRs described in this study allows the generation of large families of LRR proteins with tunable curvatures to address current challenges. The critical role of computation in the overall process is illustrated by the junction modules: both the sequences (Supplementary Fig. 5a) and the structures (Supplementary Fig. 5b) of the designed junction modules differ considerably from their closest counterparts in naturally occurring LRRs and hence could not have been obtained without energy driven design calculations. These calculations are not perfect, however, and since the small differences between the design models and the corresponding crystal structures are amplified through lever arm effects when many modules are combined, we use crystal structures of the designed building block and junction modules in the general module assembly calculations rather than the original design models.
The ability to custom design repeat proteins with well-defined shapes and curvatures has immediate application to the design of a next generation of high affinity binding proteins. Studies of native protein-protein interactions have shown that shape complementarity is a major determinant of protein binding affinity33-36. In particular, naturally occurring LRR-based binding proteins often achieve high affinity and specificity by having shapes closely conforming to the surfaces of the target proteins. The importance of this shape tuning for LRR protein molecular recognition is illustrated in Supplementary Figure 5d for the naturally occurring LRR proteins internalin A (InlA) and ribonuclease inhibitor (RI). Each protein has a curvature adapted to its target (E-cadherin and ribonuclease A, respectively) resulting in well-packed complementary protein-protein interfaces with hot-spot clusters at both the N and C termini. In contrast, swapping the respective target for each of the LRR proteins, namely InlA:ribonuclease and RI:E-cadherin complexes, results in significant clashes and large gaps.
With the capability provided by the approach described in this paper, it is now possible to design novel proteins with high backbone shape complementarity to essentially any macromolecular target of interest. Coupled with protein interface design methodology previously used to create new binding proteins based on already existing scaffolds37,38, this should allow the design of high affinity and specificity binding proteins. Such an approach complements directed evolution methods13,39,40 for obtaining high affinity binding proteins based on a single stable protein backbone which although powerful still require considerable effort. For creating a high affinity binding protein to a target of interest in the near future, a combination of our shape complementary scaffold design approach, protein-protein interface design for chemical complementarity, and limited directed evolution to optimize interactions not accurately described by computational design may prove particularly effective.
Methods
Markov transition model for natural LRR modules
To construct a Markov transition model for natural LRR modules, all set of two consecutive LRR modules were collected from the LRRML database29 and labeled based on the module length. From these data, we computed the transition probability Pa→b = Na→b/ΣiNa→i, where Na→i represents the frequency of transitions from module length a to b in the PDB. In the network model in Figure 1b, the size of a node was scaled by frequency of a module length in the PDB, and the thickness of an edge by the transition probability.
Computational design of junction modules
The initial fusion models were generated by RosettaCM24 from the motif-aligned scaffolds as described in the main text (Fig. 2a), and refined with Rosetta relax protocol with coordinate constraints41 to reduce perturbation of the structure. The fusion interface between the two heterogeneous building blocks was redesigned to improve structural compatibility using Rosetta FastRelax protocol. The protocol runs four cycles of repack, design and minimization, and during each cycle the weight for the repulsive energy term gradually increases to obtain well-packed and low-energy structure. During the design procedure, residue type constraints were added in order to favor original residue identities. After generating 1,000 design sequences, the top 10% of design sequences both by Rosetta energy and packing were retrieved and manually inspected to select the final sequences.
LRR structure modeling by iterative module assembly
Building block and junction module structures were extracted from the crystal structures of the designed LRR proteins containing one or two building block module types. Specifically, two-unit or three-unit module structures of Ncap-L22 (DLRR_A), L22-L22 (DLRR_A), Ncap-L24 (DLRR_B), L24-L24 (DLRR_B), L22→L24 (DLRR_B), L24→L22 (DLRR_E), L24→L28→L29 (DLRR_G3), L28→L29 (DLRR_G3), L29→L28 (DLRR_G3), and L24→L32→L24 (DLRR_H2) were used to elongate a LRR structure through module assembly mediated by the common flanking module. For example, module assembly of L22→L24 and L24→L22 though the common L24 unit generates the three-unit structure L22→L24→L22. The module assembly was then iteratively applied to elongate the overall structure one by one module, which resulted in the mature form of a general LRR structure. Finally, energy minimization with Rosetta was performed to eliminate potential structural defects. The crystal structure of L22→L24 was obtained from DLRR_B that has the fusion of L22-containing N-terminal capping domain (of internalin B) to L24. The L22→L28 was not used in the general module assembly due to the lack of the crystal structure.
Gene cloning, protein expression and purification
Genes encoding building block LRRs were synthesized and cloned into pET21_NESG (DLRR_A) or pET15_NESG (DLRR_B and DLRR_C) expression vectors by GeneScript. The gene fragments for each junction module were separately prepared by PCR assembly of six to eight 50–60 nucleotides oligos or by gene synthesis from Integrated DNA Technologies. Another gene fragment for the building block module to be fused was also obtained by PCR. The two gene fragments were then inserted into the plasmid of the appropriate building block protein by Gibson cloning42. The C-terminal 6xHis tag was added to all design sequences with Gly-Ser or Gly-Ser-Trp linkers where Trp was for measuring protein concentration easily.
The proteins were expressed in E. coli BL21 Star (DE3) cells at 37°C for 4 hours after induction with 0.1 mM IPTG. The cell pellets were resuspended in 20 ml of lysis buffer containing 20 mM Tris, 500mM NaCl, 30 mM imidazole, and 5% v/v glycerol (pH 8.0). Roche complete EDTA-free protease inhibitor tablet, lysozyme (1mg/ml), and DNase (1mg/ml) were also added to the lysis buffer. After sonication, the proteins were purified with a Ni-NTA column and eluted with 20 mM Tris, 500 mM NaCl, 250 mM imidazole (pH 8.0). The proteins were further purified with Superdex 200 column (GE Healthcare) equilibrated in 20 mM Tris and 50 mM NaCl at pH 8.0. The soluble expression and purity were also tested with SDS-PAGE and mass spectrometry (LCQ Fleet Ion Trap Mass Spectrometer, Thermo Scientific).
Biophysical characterization
Circular Dichroism (CD) using AVIV 62S DA spectrometer was used to investigate secondary structure contents and thermal stability. Far-UV CD spectra from 200 nm to 260 nm were measured for the protein samples in 20 mM Tris and 50 mM NaCl (pH 8.0). Thermal denaturation experiment was also performed by following the minimum at 218 nm and increasing temperature from 25°C to 90°C. Size-Exclusion Chromatography coupled to Multiple Angle Light Scattering (SEC-MALS) was performed to access oligomeric state of protein samples. Superdex 200 10/300 GL column (GE Healthcare) was equilibrated in phosphate buffered saline (PBS) buffer and used on HPLC system (LC 1200 Series, Agilent Technologies) connected to miniDAWN TREOS static light scattering detector (Wyatt Technologies). The collected data were analyzed by ASTRA software (Wyatt Technology).
X-ray crystallography
Crystals of designed LRR repeat proteins were grown by standard vapour phase diffusion methods using a TTP labtech 'Mosquito' crystallization robot with 50 nanoliter drops of protein at concentrations ranging from 15 mg/mL to 40 mg/mL equilibrated against 100 volumes of microliter individual reservoir solutions. The reservoir compositions that produced each crystal are provided in Supplementary Table 4. Crystals were then flash-cooled by rapid emersion into artificial mother liquors corresponding to the crystallization reservoir solutions supplemented with either ethylene glycol (to 25% v/v) or with PEG3350 (to 35% w/v). Diffraction data were collected on cryocooled crystals using either an in-house CCD area detector with a rotating anode x-ray generator (DLRR_A, DLRR_G3, DLRR_H2, DLRR_K) or with a CCD area detector at the Advanced Light Source X-ray synchrotron facility (DLRR_E, DLRR_I). All data were processed and scaled using program suite HKL200043. Molecular replacement was performed using program PHASER44 with computational coordinates of the individual designs produced by Rosetta as search models. Model building was performed using COOT45 and refinement was performed using program REFMAC46.
Supplementary Material
Acknowledgements
We thank J. Bolduc for data collection for DLRR_A and the members of the protein production facility at the Institute for Protein Design (Seattle, WA) for protein production. This work was supported by grants from the Defense Threat Reduction Agency (HDTRA1-11-1-0041) (to D.B.) and National Institutes of Health (R01 GM49857) (to B.L.S.).
Footnotes
Accession Codes
Atomic coordinates and structure factors have been deposited in the Protein Data Bank with the accession codes 4R58 (DLRR_A), 4R5C (DLRR_E), 4R5D (DLRR_G3), 4R6J (DLRR_H2), 4R6F (DLRR_I), and 4R6G (DLRR_K).
References
- 1.Binz HK, Amstutz P, Pluckthun A. Engineering novel binding proteins from nonimmunoglobulin domains. Nat Biotechnol. 2005;23:1257–68. doi: 10.1038/nbt1127. [DOI] [PubMed] [Google Scholar]
- 2.Skerra A. Alternative non-antibody scaffolds for molecular recognition. Curr Opin Biotechnol. 2007;18:295–304. doi: 10.1016/j.copbio.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 3.Gebauer M, Skerra A. Engineered protein scaffolds as next-generation antibody therapeutics. Curr Opin Chem Biol. 2009;13:245–55. doi: 10.1016/j.cbpa.2009.04.627. [DOI] [PubMed] [Google Scholar]
- 4.Javadi Y, Itzhaki LS. Tandem-repeat proteins: regularity plus modularity equals design-ability. Curr Opin Struct Biol. 2013;23:622–31. doi: 10.1016/j.sbi.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 5.Grove TZ, Regan L, Cortajarena AL. Nanostructured functional films from engineered repeat proteins. J R Soc Interface. 2013;10:20130051. doi: 10.1098/rsif.2013.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phillips JJ, Millership C, Main ER. Fibrous nanostructures from the self-assembly of designed repeat protein modules. Angew Chem Int Ed Engl. 2012;51:13132–5. doi: 10.1002/anie.201203795. [DOI] [PubMed] [Google Scholar]
- 7.Han SH, Lee MK, Lim YB. Bioinspired self-assembled peptide nanofibers with thermostable multivalent alpha-helices. Biomacromolecules. 2013;14:1594–9. doi: 10.1021/bm400233x. [DOI] [PubMed] [Google Scholar]
- 8.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11:725–32. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 9.Enkhbayar P, Kamiya M, Osaki M, Matsumoto T, Matsushima N. Structural principles of leucine-rich repeat (LRR) proteins. Proteins. 2004;54:394–403. doi: 10.1002/prot.10605. [DOI] [PubMed] [Google Scholar]
- 10.Kim HM, et al. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906–17. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Parker R, Mercedes-Camacho A, Grove TZ. Consensus design of a NOD receptor leucine rich repeat domain with binding affinity for a muramyl dipeptide, a bacterial cell wall fragment. Protein Sci. 2014;23:790–800. doi: 10.1002/pro.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MauLee SC, et al. Design of a binding scaffold based on variable lymphocyte receptors of jawless vertebrates by module engineering. Proc Natl Acad Sci U S A. 2012;109:3299–304. doi: 10.1073/pnas.1113193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Binz HK, et al. High-affinity binders selected from designed ankyrin repeat protein libraries. Nat Biotechnol. 2004;22:575–82. doi: 10.1038/nbt962. [DOI] [PubMed] [Google Scholar]
- 14.Forrer P, Stumpp MT, Binz HK, Pluckthun A. A novel strategy to design binding molecules harnessing the modular nature of repeat proteins. FEBS Lett. 2003;539:2–6. doi: 10.1016/s0014-5793(03)00177-7. [DOI] [PubMed] [Google Scholar]
- 15.Grove TZ, Cortajarena AL, Regan L. Ligand binding by repeat proteins: natural and designed. Curr Opin Struct Biol. 2008;18:507–15. doi: 10.1016/j.sbi.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Main ER, Xiong Y, Cocco MJ, D'Andrea L, Regan L. Design of stable alpha-helical arrays from an idealized TPR motif. Structure. 2003;11:497–508. doi: 10.1016/s0969-2126(03)00076-5. [DOI] [PubMed] [Google Scholar]
- 17.Filipovska A, Rackham O. Modular recognition of nucleic acids by PUF, TALE and PPR proteins. Mol Biosyst. 2012;8:699–708. doi: 10.1039/c2mb05392f. [DOI] [PubMed] [Google Scholar]
- 18.Reichen C, Hansen S, Pluckthun A. Modular peptide binding: from a comparison of natural binders to designed armadillo repeat proteins. J Struct Biol. 2014;185:147–62. doi: 10.1016/j.jsb.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 19.Mak AN, Bradley P, Cernadas RA, Bogdanove AJ, Stoddard BL. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science. 2012;335:716–9. doi: 10.1126/science.1216211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryou JH, Park K, Lee JJ, Kim D, Kim HS. Soluble expression of human glycoprotein Ibalpha in Escherichia coli through replacement of the N-terminal capping domain. Protein Expr Purif. 2014;101C:21–27. doi: 10.1016/j.pep.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Jung K, et al. Toll-like receptor 4 decoy, TOY, attenuates gram-negative bacterial sepsis. PLoS One. 2009;4:e7403. doi: 10.1371/journal.pone.0007403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parmeggiani F, et al. A general computational approach for repeat protein design. Journal of Molecular Biology. doi: 10.1016/j.jmb.2014.11.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang PS, et al. RosettaRemodel: a generalized framework for flexible backbone protein design. PLoS One. 2011;6:e24109. doi: 10.1371/journal.pone.0024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song Y, et al. High-resolution comparative modeling with RosettaCM. Structure. 2013;21:1735–42. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leaver-Fay A, et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–74. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheffler W, Baker D. RosettaHoles: rapid assessment of protein core packing for structure prediction, refinement, design, and validation. Protein Sci. 2009;18:229–39. doi: 10.1002/pro.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park BS, Lee JO. Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp Mol Med. 2013;45:e66. doi: 10.1038/emm.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsushima N, Miyashita H. Leucine-Rich Repeat (LRR) Domains Containing Intervening Motifs in Plants. Biomolecules. 2012;2:288–311. doi: 10.3390/biom2020288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei T, et al. LRRML: a conformational database and an XML description of leucine-rich repeats (LRRs) BMC Struct Biol. 2008;8:47. doi: 10.1186/1472-6807-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bell JK, et al. The molecular structure of the Toll-like receptor 3 ligand-binding domain. Proc Natl Acad Sci U S A. 2005;102:10976–80. doi: 10.1073/pnas.0505077102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park BS, et al. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–5. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 32.Hothorn M, et al. Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature. 2011;474:467–71. doi: 10.1038/nature10153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen R, Weng Z. A novel shape complementarity scoring function for protein-protein docking. Proteins. 2003;51:397–408. doi: 10.1002/prot.10334. [DOI] [PubMed] [Google Scholar]
- 34.Gabb HA, Jackson RM, Sternberg MJ. Modelling protein docking using shape complementarity, electrostatics and biochemical information. J Mol Biol. 1997;272:106–20. doi: 10.1006/jmbi.1997.1203. [DOI] [PubMed] [Google Scholar]
- 35.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–50. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 36.Jones S, Thornton JM. Principles of protein-protein interactions. Proc Natl Acad Sci U S A. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fleishman SJ, et al. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science. 2011;332:816–21. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Procko E, et al. Computational design of a protein-based enzyme inhibitor. J Mol Biol. 2013;425:3563–75. doi: 10.1016/j.jmb.2013.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Epa VC, et al. Structural model for the interaction of a designed Ankyrin Repeat Protein with the human epidermal growth factor receptor 2. PLoS One. 2013;8:e59163. doi: 10.1371/journal.pone.0059163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JJ, et al. A high-affinity protein binder that blocks the IL-6/STAT3 signaling pathway effectively suppresses non-small cell lung cancer. Mol Ther. 2014;22:1254–65. doi: 10.1038/mt.2014.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nivon, LG, Moretti R, Baker D. A Pareto-optimal refinement method for protein design scaffolds. PLoS One. 2013;8:e59004. doi: 10.1371/journal.pone.0059004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–5. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 43.Zbyszek Otwinowski WM. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:20. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 44.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Winn, MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–21. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.