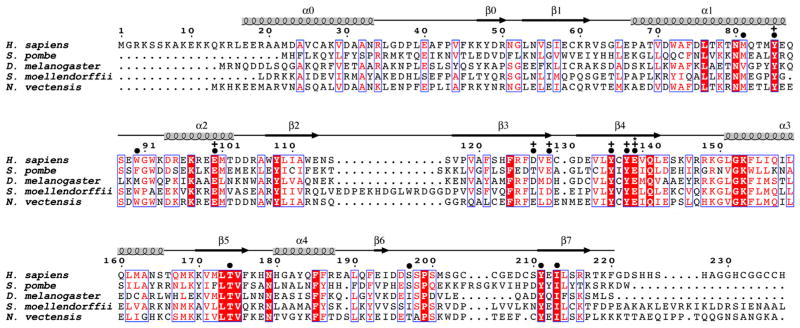
Figure 1. Sequence Alignment of NatD Orthologs.
The sequence alignment contains the following NatD orthologs: human (Homo sapiens), fission yeast (Schizosaccharomyces pombe), fruit fly (Drosophila melanogaster), spikemoss (Selaginella moellendorffii), and sea anemone (Nematostella vectensis). The blue boxes represent conserved patches of sequence alignment. Residues in red are highly conserved, and residues in white with a red background are strictly conserved. Above the sequence alignment are indicated amino acid numbering for H. sapiens and secondary structure elements. Amino acid residues are indicated that make contacts to the substrate peptide (●), show mutational sensitivity (+), or are proposed to play catalytic roles (*).