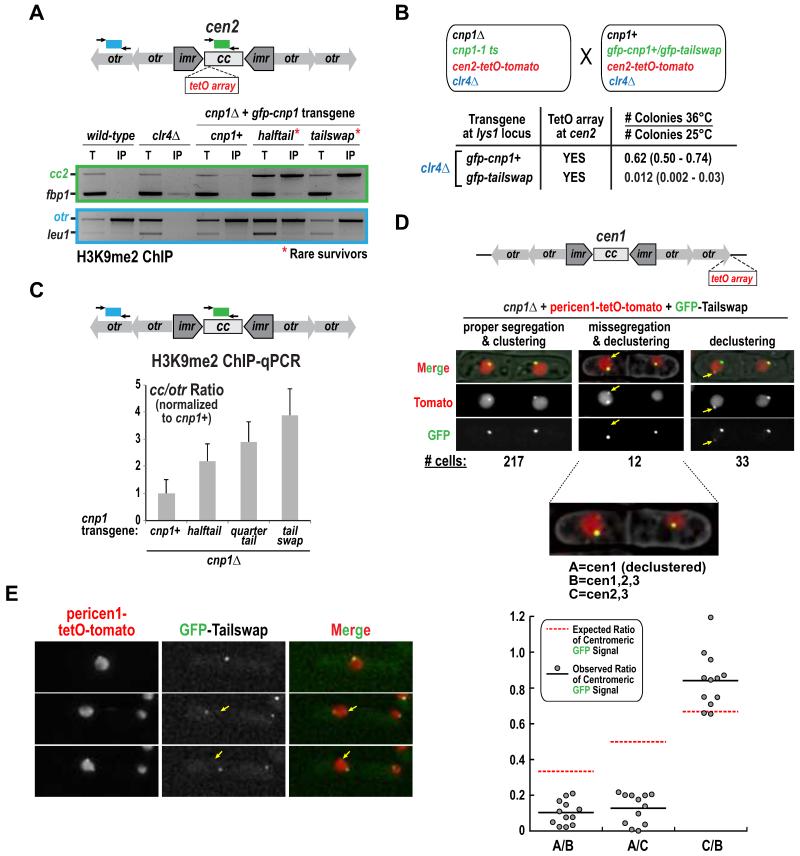
Figure 3. Evidence for centromere inactivation in Cnp1 N-tail variants in the absence of any alterations in centromeric DNA sequence.
A. H3K9me2 ChIP-PCR analysis for the indicated strains. Centromere 2 (cc2) and heterochromatic outer repeats (otr) products were compared to non-centromeric controls fbp1 and leu1, respectively. A clr4Δ control, which lacks H3K9me2, is also shown.
B. Analysis of the effect of clr4Δ on synthetic lethality of GFP-Tailswap and cen2-tetO-tomato, conducted as in Fig. 2A.
C. H3K9me2 ChIP-qPCR for the indicated strains harboring unaltered centromeres. The normalized ratio between qPCR products from Cnp1 domains (central core 1&3; cc) and heterochromatic outer repeats (otr) is shown. Error bars represent the SD (n=3). See also Fig. S3C.
D. Images of septated cells expressing GFP-Tailswap in a cnp1Δ background with a pericentromeric TetO array insertion (pericen1-tetO-tomato) on chromosome 1. The three classes of cells observed are indicated with the numbers for each class shown below the images. The yellow arrow marks the TetO-tomato focus that is declustered and has low GFP signal. In cells with missegregation and declustering, GFP intensity was measured at each red (TetO-Tomato) focus and the indicated ratios are plotted below. The expected ratio, assuming equal amount of GFP-Cnp1 is present at each centromere, is shown with a red dashed line. Scale bars are 3 μm.
E. Representative time-lapse images providing evidence for centromere inactivation. Arrow indicates a TetO-tomato focus that has very low GFP signal. Scale bar is 3 μm.