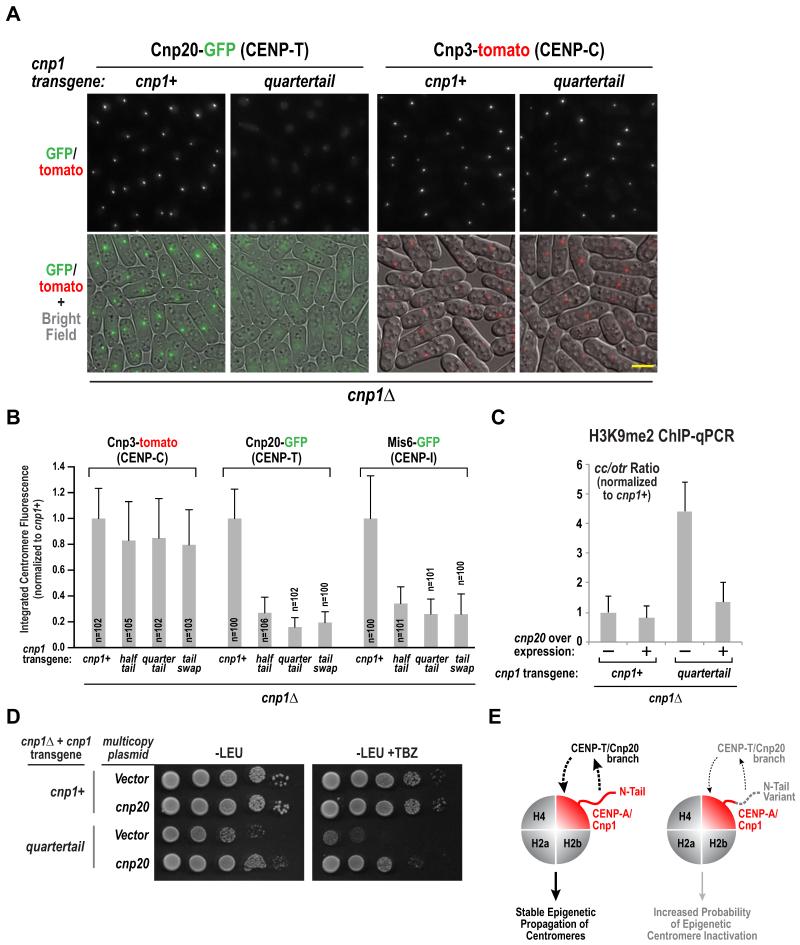
Figure 4. Cnp1 N-tail variants selectively reduce centromeric accumulation of the Cnp20/CENP-T branch of the CCAN.
A. Representative images of Cnp20-GFP and Cnp3-tomato in the indicated strains. Scale bar represents 5 μm. See also Fig. S4A.
B. Integrated fluorescence intensity of Cnp20-GFP, Mis6-GFP and Cnp3-tomato foci was measured and plotted for the indicate strains as in Fig. 1F&G. Error bars represent the standard deviation.
C. ChIP-qPCR with H3K9me2 antibody for the indicated conditions. The normalized ratio of products from Cnp1-enriched regions (cc) and heterochromatic outer repeats (otr) is displayed. See also Fig. S4B. Error bars represent the SD (n=3).
D. Serial dilutions of indicated strains harboring empty or Cnp20-overexpressing multicopy pREP1 plasmid were plated on minimal medium lacking leucine (PMG –LEU), with or without 15 μg/ml TBZ, and grown at 33°C. A ten-fold dilution series is shown for each strain.
E. Schematic summary of key findings. The Cnp1/CENP-A N-tail is required to recruit the Cnp20/CENP-T branch of the CCAN, which in turn is required for stable epigenetic propagation of centromeres (left). Cnp1 N-tail variants are normally loaded and support outer kinetochore assembly but the CENP-T branch of the CCAN is selectively reduced, increasing the probability of centromere inactivation and chromosome missegregation (right).