Background: Mitotic SUMOylation is critical for faithful chromosome segregation in eukaryotes.
Results: PICH (Plk1-interacting checkpoint helicase) is a SUMO-interacting protein and a mitotic SUMO substrate.
Conclusion: PICH can be regulated by binding to SUMOylated proteins and through its own SUMOylation at mitotic centromeres.
Significance: The regulation of PICH by SUMO interaction and SUMOylation reveals a novel role for mitotic SUMOylation in centromeric chromatin organization.
Keywords: Centromere, Mitosis, SUMO-interacting Motif (SIM), Sumoylation, Xenopus, PICH, SUMO
Abstract
Mitotic SUMOylation has an essential role in faithful chromosome segregation in eukaryotes, although its molecular consequences are not yet fully understood. In Xenopus egg extract assays, we showed that poly(ADP-ribose) polymerase 1 (PARP1) is modified by SUMO2/3 at mitotic centromeres and that its enzymatic activity could be regulated by SUMOylation. To determine the molecular consequence of mitotic SUMOylation, we analyzed SUMOylated PARP1-specific binding proteins. We identified Polo-like kinase 1-interacting checkpoint helicase (PICH) as an interaction partner of SUMOylated PARP1 in Xenopus egg extract. Interestingly, PICH also bound to SUMOylated topoisomerase IIα (TopoIIα), a major centromeric small ubiquitin-like modifier (SUMO) substrate. Purified recombinant human PICH interacted with SUMOylated substrates, indicating that PICH directly interacts with SUMO, and this interaction is conserved among species. Further analysis of mitotic chromosomes revealed that PICH localized to the centromere independent of mitotic SUMOylation. Additionally, we found that PICH is modified by SUMO2/3 on mitotic chromosomes and in vitro. PICH SUMOylation is highly dependent on protein inhibitor of activated STAT, PIASy, consistent with other mitotic chromosomal SUMO substrates. Finally, the SUMOylation of PICH significantly reduced its DNA binding capability, indicating that SUMOylation might regulate its DNA-dependent ATPase activity. Collectively, our findings suggest a novel SUMO-mediated regulation of the function of PICH at mitotic centromeres.
Introduction
SUMOylation is a posttranslational modification that leads to diverse biological consequences. The addition of small ubiquitin-like modifier (SUMO)3 affects target proteins by altering their subcellular localization, enzymatic activity, and/or protein-protein/protein-DNA interactions (1–5). Similar to ubiquitylation, SUMO conjugation to its substrates requires a cascade of three enzymes: an activating E1 enzyme, a conjugating E2 enzyme, and typically a SUMO E3 ligase enzyme. All SUMO modifications utilize the same E1 (Uba2/Aos1) and E2 (Ubc9) enzymes, but different E3 enzymes have been identified that show specificity to certain targets (6). Multiple lines of evidence show that SUMOylation is a vital posttranslational modification to ensure the proper segregation of chromosomes during mitosis (7, 8). Consistent with observed segregation defects, many mitotic proteins have been identified as SUMO targets (7).
On mitotic chromosomes, a major SUMO signal is observed at the centromeric region in both somatic cells and Xenopus egg extract (XEE) cell-free assay (9, 10). Using the XEE assays, we have previously identified two major PIASy-dependent mitotic chromosomal SUMO2/3 substrates: DNA topoisomerase IIα (TopoIIα) and poly(ADP-ribose) polymerase 1 (PARP1) (11, 12). TopoIIα was one of the first mitotic SUMOylated substrates identified in budding yeast and vertebrates (11, 13) and is pivotal for DNA decatenation to separate sister chromatids during chromosome segregation. Accumulating evidence indicates that SUMOylation is important for the regulation of TopoIIα activity (14, 15). Another robust mitotic SUMOylation substrate, PARP1 (12), is a member of the PARP family that catalyzes the formation of poly(ADP-ribose) on target proteins, leading to multifaceted biological consequences (16). Although we have previously shown potential PARP1 activity regulation by SUMOylation on mitotic chromosomes (12), the comprehensive mitotic role of PARP1, as well as how SUMO modification affects the function of PARP1 during mitosis, has not yet been determined.
SUMO modification often provides a new site for protein-protein interactions (17–19), and non-covalent interactions between SUMO-interacting motif (SIM)-containing proteins and SUMOylated proteins have been shown to produce multiple critical functional consequences (20–22). To extend our understanding of the downstream effects of SUMOylation at mitotic centromeres, we intended to identify SUMOylation-dependent binding protein(s) using PARP1 as bait. We identified Polo-like kinase 1 (Plk1)-interacting checkpoint helicase (PICH), which is also known as ERCC6-like protein and belongs to the SNF2 family of ATPases, as a novel SUMO-interacting partner. Prior studies have shown that PICH is essential for the proper segregation of chromosomes during mitosis (23–25). In this study, we detected PICH as a novel SUMO substrate on mitotic chromosomes. In vitro SUMOylated PICH showed reduced DNA binding capability, implicating the SUMO-dependent regulation of PICH activity. Altogether, we propose a novel regulation of PICH function at mitotic centromeres by SUMOylation.
EXPERIMENTAL PROCEDURES
Plasmids and Antibody Preparation
Human PICH (PICHhs) cDNA was amplified from a plasmid obtained from Addgene (plasmid 41163: Nigg CB62) (23) and subcloned into pPIC3.5K fused to calmodulin-binding protein and with a T7 tag (14). PICHhs cDNA for mRNA expression was cloned into the pTGFC70 plasmid, a generous gift from Dr. Funabiki, and utilized for mRNA expression as described previously (26). Partial cDNAs for Xenopus laevis PICH were obtained by PCR amplification from X. laevis cDNA based on expressed sequence tag clone sequences that are homologous to PICHhs. The obtained partial PICHxl cDNAs were subcloned into pET28a and pMalc5x for recombinant protein expression.
A polyclonal antibody against PICHxl was generated in rabbits by injecting His6-tagged recombinant PICHxl fragments (Pacific Immunology, Ramona, CA), and the specific antibody was purified via maltose-binding protein (MBP)-tagged PICHxl affinity column chromatography (11). A guinea pig anti-SUMO2/3 antibody and chicken anti-CENPA antibody were prepared as described previously (12). Commercial antibodies used in this study were S-protein-HRP and anti-T7-HRP (EMD Millipore, Billerica, MA), monoclonal anti-GFP (JL-8) (Clontech), monoclonal anti-histone 2B (Abcam, Cambridge, MA), monoclonal anti-PAR (Trevigen, Gaithersburg, MD), and fluorescently labeled secondary antibodies (Life Technologies).
Xenopus Egg Extract Immunofluorescence and Immunoblotting
Low-speed extracts arrested in metaphase by cytostatic factor (CSF) from X. laevis egg and sperm nuclei were prepared using standard protocols (27). An interphase extract was obtained by releasing CSF upon the addition of CaCl2 to the CSF extracts (27). The mitotic chromosomes used for the immunofluorescence analysis were prepared as described previously (9). Mitotic SUMOylation was inhibited by the addition of a dominant-negative form of Ubc9 (dnUbc9) at a concentration of 150 ng/μl before the induction of mitosis. The localization of human PICH on mitotic chromosomes was observed by GFP signals from exogenously expressed human PICH-EGFP mRNA in the extract (26). The DNA was stained with Hoechst 33342 dye (EMD Millipore), and the samples were mounted using VECTASHIELD (Vector Laboratory). The specimens were analyzed using a Nikon TE2000-U microscope with a Plan Apo 100×/1.40 objective, and the images were taken with a Retiga SRV CCD camera (QImaging) using the Volocity imaging software (Improvision).
Chromatin isolation for immunoblotting was performed as described previously (11). Recombinant SUMO2GG-EGFP protein (100 ng/μl extract) was added for the supershift assay shown in Fig. 2d. The samples were subjected to immunoblotting with the indicated antibodies.
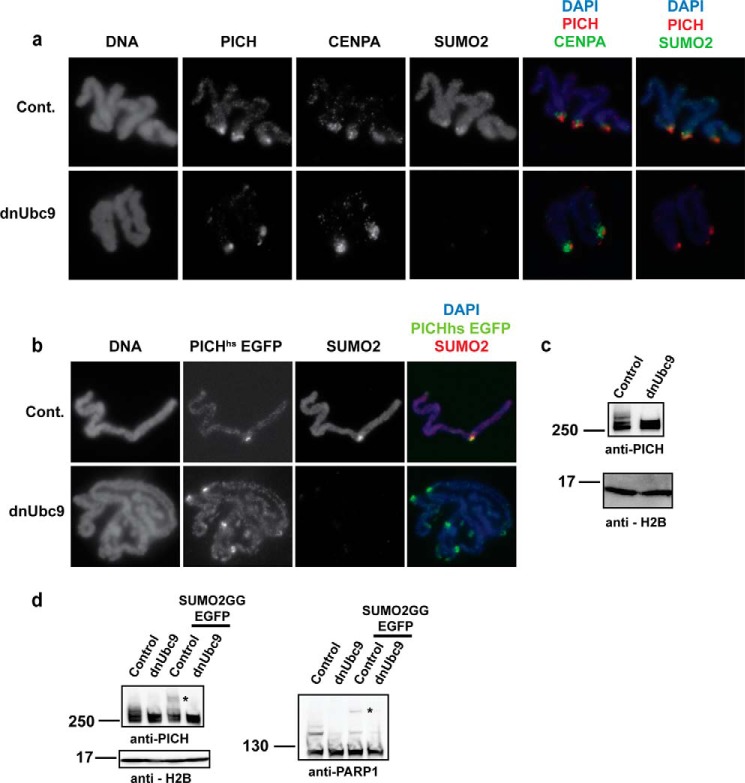
FIGURE 2.
PICH is enriched at the centromere independently of mitotic SUMOylation and is SUMOylated on mitotic chromosomes. a, PICH is enriched at the centromere, co-localizing with CENPA and SUMO2/3, and the inhibition of SUMOylation by dnUbc9 does not alter PICH localization at the mitotic chromosomes. Replicated mitotic chromosomes were isolated from XEE and subjected to immunofluorescence staining with the indicated antibodies. Cont., control. b, PICHhs was enriched at the centromere, similar to endogenous PICH on mitotic chromosomes isolated from XEE. PICHhs-EGFP mRNA was added to XEE with and without the addition of dnUbc9 and was visualized on mitotic chromosomes along with CENPA and SUMO2/3. c, PICH is posttranslationally modified on mitotic chromosomes in XEE. Non-replicated mitotic chromosomes were isolated from XEE and analyzed by immunoblotting with an anti-PICH antibody. SUMOylation was inhibited by the addition of dnUbc9. d, PICH is SUMOylated on mitotic chromosomes in XEE. SUMO2GG-EGFP was added to the XEE, and non-replicated mitotic chromosomes were isolated and analyzed by immunoblotting. PARP1, a known SUMO substrate, served as a positive control for the PARP1-SUMO-EGFP supershift (indicated with asterisks).
Pulldown Assay
Pulldown assays were performed as described previously, with a few modifications (28). Non-SUMOylated and SUMOylated bait proteins were bound to either anti-T7-agarose or S-protein-agarose (EMD Millipore), depending on the affinity tag fused to the bait protein. XEE was diluted three times with immunoprecipitation buffer (20 mm NaPi, pH 7.8, 18 mm β-glycerol phosphate, pH 7.5, 5 mm MgCl2, 100 mm NaCl, 1 mm CaCl2, and 10 μm ZnCl2) and clarified by centrifugation at 25,000 × g for 30 min at 4 °C. The soluble fraction was further diluted with an equal volume of ChIP buffer (immunoprecipitation buffer containing 0.2% Triton X-100 and 0.2% Tween 20) and then incubated with protein-bound beads for 1 h at 4 °C. SENP2-CD digestion was performed by incubating ∼50 ng/μl SENP2-CD with beads in ChIP buffer for 30 min at 25 °C. Half of the volume of 3× SDS-PAGE sample buffer was added to the SENP2-CD-digested samples, resolved by SDS-PAGE on 8–16% gradient gels (Life Technologies), and analyzed by silver staining and/or immunoblotting, as indicated. For the pulldown analysis of PICHhs, either the egg extract expressing full-length PICHhs-EGFP or 15 μg of purified PICHhs in ChIP buffer containing 5% gelatin was incubated with the bait. For SUMO2 pulldown, GFP-SUMO2 and GFP-SUMO2 × 4 (GFP-SUMO chain) were incubated with GFP trap magnetic beads (Bulldog Bio Inc.). The pulled down samples were analyzed as described above using silver staining and immunoblotting, as indicated.
Protein Purification and in Vitro SUMOylation Assays
Recombinant PICHhs with calmodulin-binding protein (CBP) and T7 tags was purified from a GS115 strain of Pichia pastoris as described previously (14). The E1 complex (Aos1/Uba2 heterodimer), Ubc9, dnUbc9, PIASy, and SUMO2 were purified as described previously (12, 14).
In vitro SUMOylation assays were performed with 40 nm E1, 80 nm Ubc9, 40 nm PIASy, and 3 μm PARP1 N650 and SUMO2 (6–10× substrate concentration), unless otherwise indicated, for ∼2 h at 25 °C. SUMOylated PICH and PARP1 were analyzed by immunoblotting using a T7-HRP antibody.
Electrophoretic Mobility Shift Assay
Non-SUMOylated PICHhs was prepared via in vitro SUMOylation with SUMO2-G, a non-conjugatable form of SUMO2. Linearized pBluescript DNA (∼25 nm or 50 nm) was incubated in a 10-μl in vitro SUMOylation reaction containing PICHhs (∼1 μm) for 20 min on ice. The entire reaction was separated on a 0.8% agarose gel in pre-chilled buffer at 25 V for 3 h. The DNA was visualized via ethidium bromide staining (post run). Quantification of the non-shifted DNA was performed using the ImageJ and KaleidaGraph software.
PARylation Assay
The pulled down fractions with non-SUMOylated and SUMOylated PARP1 were used for a PARylation assay, as described previously (12). The beads were incubated with SUMOylation buffer containing NAD+, and the samples were subjected to an immunoblotting analysis of PARylation with an anti-PAR antibody.
RESULTS
SUMOylation of PARP1 Mediates Protein-Protein Interactions
We previously showed that PARP1 SUMOylation does not alter its enzymatic activity but does result in subtle differences in the PARylation profile of mitotic chromosomal proteins (12). To examine whether the SUMOylation of PARP1 could affect its substrate specificity, we performed a PARylation reaction with a PARP1 pulldown fraction using XEE. Either SUMOylated or non-SUMOylated PARP1 was captured on agarose beads, and the beads were then incubated with XEE to isolate binding proteins. The beads were incubated in a reaction buffer containing NAD+ to initiate PARylation by PARP1. Consistent with our previous finding, PARP1 showed robust self-PARylation activity regardless of SUMOylation (Fig. 1a, lanes 1 and 2); however, we observed a difference in the PARylation profile when we compared PARylation on beads between samples without XEE and samples after pulldown with XEE using PARP1 and SUMOylated PARP1 (Fig. 1a, lanes 3 and 4). In the pulldown samples incubated with XEE, there was an increase in the PARylation of ∼150 kDa for non-SUMOylated PARP1, whereas more PARylation, ∼250 kDa, was observed for SUMOylated PARP1 (Fig. 1a). This finding suggests that PARP1 promotes the PARylation of its interacting proteins and that the SUMOylation of PARP1 could alter its substrate specificity in XEE. Conjugation of SUMO to cellular proteins is known to create new docking sites for protein-protein interactions through a SIM (29). Our results are consistent with this notion because SUMOylation potentially regulates the interacting proteins of PARP1. To examine this further, we sought to identify the binding proteins for PARP1 and SUMOylated PARP1. We utilized a PARP1 N-terminal 650-amino acid fragment (PARP1-N650) that contains most of the SUMOylation sites that we identified by in vitro SUMOylation with a series of mutants (data not shown). Because the expected binding proteins will be larger than 100 kDa, the use of PARP1-N650, the molecular mass of which is ∼85 kDa with the affinity tag, will be beneficial for distinguishing the bands of the bound proteins by SDS-PAGE. In addition, we introduced SENP2 digestion after pulldown to eliminate the bands of SUMOylated bait for better visibility of the bound proteins (Fig. 1b). As shown in Fig. 1c, PARP1 bound to many proteins in XEE, and several proteins were specific for SUMOylated PARP1, suggesting that PARP1 SUMOylation mediated specific protein-protein interactions.
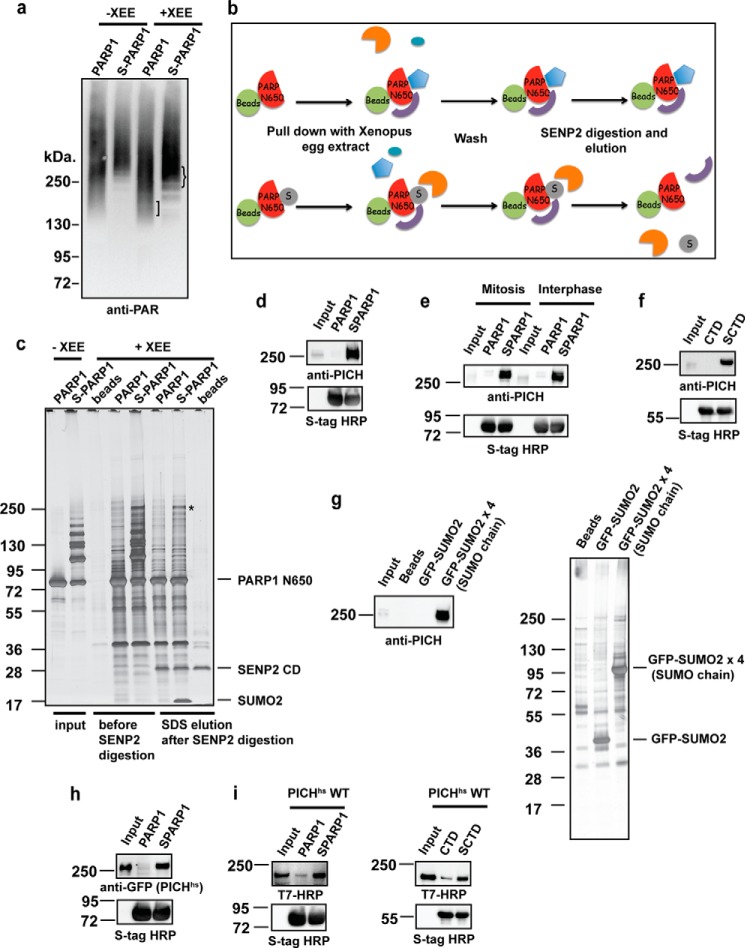
FIGURE 1.
PICH (also known as ERCC6-like) interacts with SUMOylated substrates. a, differences in the PARylation profiles of PARP1 (PARP1) and SUMOylated PARP1 (S-PARP1) pulled down samples from XEE. The PARylation profile of pulled down samples was analyzed as described under “Experimental Procedures.” The baits are indicated on each lane, and lanes 1 and 2 were not incubated with XEE, showing the self-PARylation of PARP1 and S-PARP1, respectively. b, schematic diagram of the pulldown assay. PARP1/TopoIIα-CTD and SUMOylated PARP1/TopoIIα-CTD (SCTD) were bound to S-protein-agarose beads. After incubation with XEE and washes, the bound proteins were incubated with the catalytic domain of SENP2. SENP2 assisted in the elimination of SUMOylated PARP1/TopoIIα-CTD bands during the analysis by deconjugating SUMO. c, isolation of SUMOylated PARP1-specific binding proteins. A pulldown assay was performed with the non-SUMOylated (PARP1) and SUMOylated PARP1-N650 (S-PARP1) fragment, which were bound to S-agarose beads. Bound proteins were visualized by silver staining. SENP2 digestion eliminated the SUMOylated PARP1-N650 bands. A 250-kDa protein that specifically interacted with S-PARP1 (indicated by an asterisk) was identified as PICH by LC-MS/MS. d, specific interaction of PICH with SUMOylated PARP1-N650. Pulldown fractions digested by SENP2 as in c were subjected to immunoblotting using an affinity-purified anti-PICH antibody. The CSF input lane on the membrane was ∼3% of the XEE used for the assay. The amount of bait (PARP1-N650) was analyzed by immunoblotting using S-tag HRP. e, cell cycle-independent PICH interaction with SUMOylated substrates. PICH interaction with SUMOylated PARP1-N650 was analyzed by pulldown assays using mitotic or interphase XEE. S-tag HRP was used to detect the amount of bait. f, PICH interacts with multiple SUMOylated substrates. Pulldown assays were performed with non-SUMOylated and the SUMOylated TopoIIα CTD (SCTD). SENP2 eluted fractions were analyzed by immunoblotting using an anti-PICH antibody. S-tag HRP was used as the bait loading control after SENP2 digestion. g, PICH interacts with SUMO chain. A pulldown assay was performed with GFP-SUMO and GFP-SUMO chain. The pulled down fractions were analyzed by silver staining and immunoblotting using an anti-PICH antibody. h, PICH-SUMO interaction is conserved in humans. XEE expressing EGFP-tagged PICHhs was subjected to pulldown assays. PICHhs in pulldown fractions was analyzed by immunoblotting with an anti-GFP antibody. S-tag HRP depicted the amount of bait in each sample. I, PICHhs directly interacts with SUMOylated PARP1-N650 and CTD. Purified recombinant PICHhs fused to a T7 tag was incubated with non-SUMOylated and SUMOylated PARP1-N650 or TopoIIα-CTD. The bound fraction was analyzed by immunoblotting using an anti-T7 tag-HRP antibody.
PICH Interacts with Multiple SUMOylated Substrates and SUMO Chain
The prominent 250-kDa band present only in the SUMOylated PARP1-N650 pulldown fraction (Fig. 1c, asterisk) was subjected to LC-MS/MS analysis and identified as ERCC6-like protein, also known as PICH. To confirm the LC-MS/MS result, we prepared an anti-PICH antibody with Xenopus laevis PICH fragments. Full-length Xenopus laevis PICH cDNA information is not available in databases, but we found expressed sequence tag clones that show homology to human PICH. Thus, we isolated these clones from Xenopus cDNA and then prepared proteins for antigens. Thus far, we have been unable to obtain a full-length Xenopus laevis PICH cDNA using PCR-based cloning. Immunoblotting of the pulldown fractions with a PICH-specific antibody confirmed that PICH specifically interacts with SUMOylated PARP1-N650 (Fig. 1d). To further characterize the PICH/SUMO interaction, we examined whether the interaction could be regulated by cell cycle stages as well as its specificity for SUMOylated proteins. Pulldown assays performed with mitotic and interphase extracts indicated that PICH interacted with SUMOylated PARP1-N650, irrespective of the cell cycle stage (Fig. 1e). Additionally, PICH bound to a SUMOylated TopoIIα fragment, another major SUMOylated protein at mitotic chromosomes (Fig. 1f) (11, 13). PICH also bound to SUMO chain and not to a single SUMO moiety, suggesting the presence of potentially multiple SIMs in PICH (Fig. 1g). These results suggest that PICH interacts with the SUMO moiety, regardless of the cell cycle stage and species of protein that is SUMOylated. To confirm whether the PICH/SUMO interaction is conserved for PICHhs, we performed a pulldown assay with XEE expressing PICHhs. Consistent with endogenous PICH, PICHhs specifically bound to SUMOylated PARP1, indicating that the PICH/SUMO interaction is conserved between Xenopus and humans (Fig. 1h). Because PICH forms a complex with other proteins (30–32), it is possible that PICH interacts with SUMOylated PARP1 through its binding partner(s). To examine whether PICH directly binds to SUMOylated proteins, we prepared a recombinant PICHhs protein by expressing it in yeast, P. pastoris. Pulldown assays were performed using PARP1-N650 and the TopoIIα fragment as baits. As shown in Fig. 1i, recombinant PICHhs clearly bound more highly to the SUMOylated baits than to the non-SUMOylated baits, indicating that PICH could directly bind to SUMOylated substrates and, thus, could have a SIM in its primary sequence. Supporting this observation, computational prediction indicated multiple potential SIMs in the PICHhs primary sequence.
PICH Is Enriched at the Centromere of Mitotic Chromosomes, and Its Localization Is Not Dependent on SUMOylation
Mitotic chromosomal SUMOylation is mainly detected at the centromere. We have recently observed that some centromeric proteins not only interact with SUMO-modified TopoIIα but also require mitotic SUMOylation for their proper localization.4 PICH was shown to localize at mitotic centromeres in mammalian cells (23, 33), indicating that its localization might be dependent on SUMOylation. To test this possibility, condensed replicated chromosomes were isolated in the presence and absence of mitotic SUMOylation and were subjected to immunofluorescence analysis using an anti-PICH antibody. Endogenous PICH was enriched at the centromere of replicated mitotic chromosomes, co-localizing with CENPA, a centromere-specific H3 variant, and SUMO2/3. This enrichment was not altered when SUMOylation was inhibited by the addition of a dominant-negative form of Ubc9 (Fig. 2a). The localization of EGFP-tagged PICHhs was similar to endogenous PICH and was not altered upon the inhibition of SUMOylation (Fig. 2b). To bolster our observation that SUMOylation does not alter PICH localization on mitotic chromosomes, the amount of endogenous PICH was analyzed by immunoblotting. Mitotic chromosomes were isolated from XEE, and the amount of PICH on chromosomes with and without SUMOylation was evaluated using an anti-PICH antibody. Consistent with the immunofluorescence data, the overall amount of PICH was not altered, but surprisingly, we observed a shift above the expected molecular weight of PICH. The shift was absent on the chromosomes isolated under SUMOylation inhibition by dnUbc9 (Fig. 2c). This finding suggests that PICH is most likely posttranslationally modified in a SUMO-dependent manner on mitotic chromosomes. To test whether the posttranslational modification was SUMOylation itself, we added EGFP (∼25 KDa)-tagged SUMO2GG to the mitotic extract and assessed supershifted PICH on mitotic chromosomes. A fraction of PICH indeed was supershifted with the addition of EGFP-SUMO2GG, similar to a previously identified SUMO substrate, PARP1 (12). This shift was not observed on chromosomes isolated from extracts with dnUbc9 (Fig. 2d). Altogether, we concluded that PICH is a SUMO substrate on mitotic chromosomes.
PICHhs Is a Strong PIASy-dependent SUMOylation Substrate in Vitro
To examine whether PICH can be SUMOylated in vitro, similar to our previously identified substrates, TopoIIα and PARP1 (12, 14), we performed an in vitro SUMOylation assay with recombinant PICHhs as a substrate. In contrast to PARP1, PICHhs could not be SUMOylated at the highest Ubc9 concentration (600 nm) tested, at which PARP1 could be modified by SUMO2 (Fig. 3a) (12). PICH modification by SUMO2, however, was detected in the reaction with the lowest concentration of PIASy (10 nm) (Fig. 3b), a major mitotic SUMO E3 ligase. Most SUMO substrates, including TopoIIα and PARP1, can be SUMOylated in the absence of the E3 ligase in an in vitro assay, albeit the efficiency of the modification is much lower than the reaction with E3 ligases. Within this context, PICH is a unique SUMO substrate that eminently requires an E3 ligase for its modification in vitro.
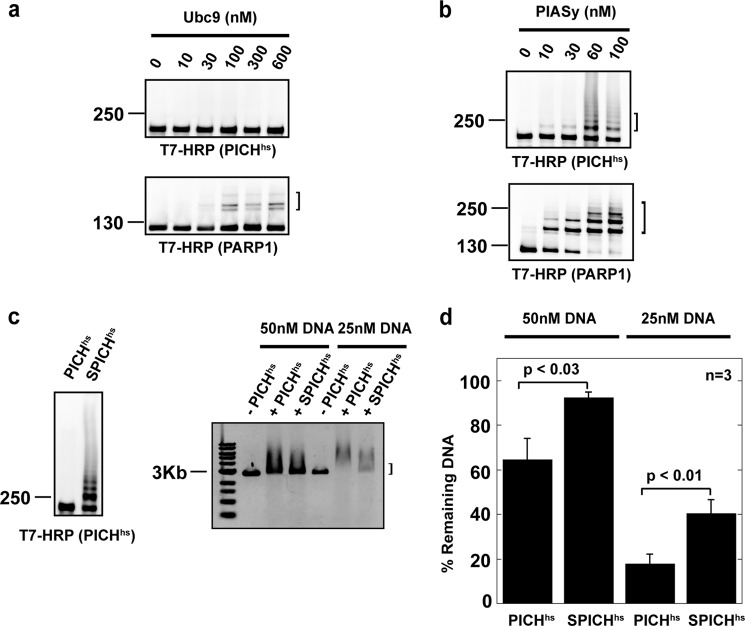
FIGURE 3.
PICHhs is SUMOylated in vitro in a PIASy-dependent manner, and PICHhs SUMOylation reduces DNA binding in vitro. a and b, in vitro SUMOylation reactions were performed as described previously (12, 14) with 40 nm E1, 80 nm Ubc9, 40 nm PIASy, 0.5 μm PICHhs or PAPR1, and 10 μm SUMO2, unless otherwise indicated, and were analyzed by immunoblotting using an anti-T7 antibody conjugated to HRP. PIASy is required for robust PICH SUMOylation. SUMOylation reactions with increasing concentrations of Ubc9 in the absence of PIASy (a) and the reaction with increasing PIASy are shown (b). Brackets indicate SUMOylated species. c, effect of PICH SUMOylation on PICH/DNA interaction. PICHhs was SUMOylated (SPICHhs) in vitro and incubated with linear pBluescript plasmid. PICHhs-DNA interactions were observed using agarose gels, followed by EtBr staining. The bracket indicates the non-shifted DNA position. d, quantification of the percentage of non-shifted DNA in the EMSA. The p values and S.D. were calculated from three independent experiments.
SUMOylation of PICHhs Alters Its Ability to Bind to DNA in Vitro
Because PICH is suggested to play a role in centromeric nucleosome eviction during mitosis, we sought to examine whether PICH SUMOylation has any impact on its enzymatic activity (30). Because PICH has been previously characterized as a DNA-dependent ATPase (33), we examined the effect of PICH SUMOylation on the PICH/DNA interaction. PICHhs was SUMOylated in vitro (Fig. 3c), and its ability to bind DNA was observed by an electrophoretic mobility shift assay by adding DNA to the in vitro SUMOylation reaction. The mobility of the DNA was significantly reduced when PICHhs was SUMOylated in comparison with non-SUMOylated PICHhs (Fig. 3c). We still observed a modest shift in the DNA in the lane where PICHhs was subjected to SUMOylation (Fig. 3c); however, this result was expected because we could not obtain more than 50% SUMOylated PICHhs in the reactions (Fig. 3c). Nevertheless, the quantification of the remaining DNA with or without SUMOylation from three independent experiments indicated that the DNA/PICHhs interaction is negatively affected by more than 20% when PICHhs is SUMOylated (Fig. 3d). This result suggests that the SUMOylation of PICH might impair its DNA-dependent ATPase activity. Collectively, our findings suggest that the SUMOylation of PICH at mitotic centromeres could regulate its activity during mitosis.
DISCUSSION
Our results imply that two mechanisms of the SUMOylation-dependent regulation of PICH function at the mitotic centromere: 1) the SUMOylation of PICH, which could inhibit the PICH/DNA interaction; and 2) the interaction of SUMOylated centromeric proteins (PARP1 and TopoIIα) through the SIM of PICH. This novel regulation of PICH function by mitotic SUMOylation might affect the regulation of centromeric nucleosomes by PICH, which has been shown to be important for resolving sister chromatids during anaphase.
Inhibition of PICH Activity by SUMOylation
The inhibition of DNA binding by the SUMOylation of PICH could be the result of 1) a direct effect on the DNA binding site of PICH by SUMOylation or 2) a conformational change by intramolecular interactions of SUMO and SIM. In the first case, we expect that the SUMOylation sites of PICH could be close to its DNA binding site, which is currently not defined. Because PICH SUMOylation could not be observed in the Ubc9-dependent reaction, the SUMOylation sites might not be a canonical SUMOylation sequence that can be mediated by direct interaction with the Ubc9-SUMO adduct (29, 34). The identification of the SUMOylation sites and the SIM of PICH will further help us uncover these questions.
Interaction of PICH with Other SUMOylated Proteins
We initially identified PICH as one of the binding proteins of SUMOylated PARP1 in XEE, but further analyses showed that PICH also bound to SUMOylated TopoIIα C-terminal domain (CTD) and SUMO chain (Fig. 1, d, f, and g). This finding suggests that PICH can bind non-specifically to SUMOylated proteins. The DNA helicase mutated in Bloom syndrome, BLM, a known PICH-binding protein, is SUMOylated in cells (35). Thus, it is possible that the SUMOylation of BLM could modulate BLM-PICH complex function via an interaction between the SUMO moiety on BLM and the SIM of PICH. Another piece of intriguing evidence could be in relation to the SUMOylation of TopoIIα. The inhibition of TopoIIα activity by ICRF-193 treatment has been shown to increase PICH-loaded ultrafine DNA bridges (36). Interestingly, the same inhibitor is known to increase TopoIIα SUMOylation in mitosis (37). Thus, it is possible that the hyper-SUMOylation of TopoIIα by ICRF-193 contributes to the retention of PICH at ultrafine DNA bridges during anaphase, in addition to the maintenance of catenated DNA via the inhibition of TopoIIα activity. The possibility remains that our initial hypothesis of PICH being a potential PARP1 substrate for the PARylation of SUMOylated PARP1 is true. To date, we have not been able to detect the PARylation of PICH in an in vitro assay, although this hypothesis could hold true for mitotic chromosomes.
Implication of SUMO-dependent Regulation of PICH for Mitotic Centromere Organization
The ATPase activity of PICH is potentially required for centromeric nucleosome eviction (30); therefore, the SUMOylation of PICH could attenuate PICH-dependent centromeric nucleosome eviction by inhibiting the DNA-dependent ATPase activity of PICH. Within this context, because the inhibition of PICH SUMOylation could increase the activity of PICH at mitotic centromeres, fewer centromeric histones are expected when PICH SUMOylation is inhibited. Supporting that notion, a previous study indicated that the elimination of PICH in cells increases centromeric histones (30). Recent in vitro data with PICH did not show the efficient remodeling activity of nucleosomes with canonical histones or octamers composed of the centromeric histone variant CENPA (38), indicating an unidentified histone specificity for PICH nucleosome eviction function at centromeres. Combining PICH's capability of interacting with SUMO and considering the potential SUMOylation of histones at heterochromatin loci (39), it is possible that the nucleosome remodeling activity of PICH is more effective toward SUMOylated histones. Future studies using potential SUMOylation-deficient mutants and SUMO interaction-deficient mutants in XEE and somatic cells will provide insight into the precise role of the SUMOylation-dependent regulation by PICH of chromatin organization at mitotic centromeres.
Acknowledgments
We thank H. Funabiki for sharing the pTGFC70 plasmid and A. Arnaoutouv for SUMO chain plasmid. We also thank NPG Language Editing for editorial assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant GM80278 through the NIGMS (to Y. A.) and is currently supported by bridge funding from the University of Kansas and in part by a general research fund (Grant 2301743) from the University of Kansas.
H. Ryu, M. Yoshida, A. Kumagai, W. G. Dunphy, M. Dasso, and Y. Azuma, manuscript in preparation.
- SUMO
- small ubiquitin-like modifier
- SIM
- SUMO-interacting motif
- CSF
- cytostatic factor
- CTD
- C-terminal domain
- SENP2-CD
- sentrin-specific protease 2 catalytic domain
- PICH
- Plk1-interacting checkpoint helicase
- Plk1
- Polo-like kinase 1
- PICHhs
- human PICH
- PICHxl
- X. laevis PICH
- XEE
- Xenopus egg extract
- PARP1
- poly(ADP-ribose) polymerase I
- PAR
- poly(ADP-ribose)
- dnUbc9
- dominant-negative Ubc9
- TopoIIα
- topoisomerase IIα
- EGFP
- enhanced GFP
- S-tag HRP
- S-protein-HRP
- CENPA
- centromere protein A
- PIASy
- protein inhibitor of activated STAT.
REFERENCES
- 1. Goodson M. L., Hong Y., Rogers R., Matunis M. J., Park-Sarge O. K., Sarge K. D. (2001) SUMO-1 modification regulates the DNA binding activity of heat shock transcription factor 2, a promyelocytic leukemia nuclear body associated transcription factor. J. Biol. Chem. 276, 18513–18518 [DOI] [PubMed] [Google Scholar]
- 2. Hong Y., Rogers R., Matunis M. J., Mayhew C. N., Goodson M. L., Park-Sarge O. K., Sarge K. D. (2001) Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J. Biol. Chem. 276, 40263–40267 [DOI] [PubMed] [Google Scholar]
- 3. Matunis M. J., Wu J., Blobel G. (1998) SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol. 140, 499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pichler A., Melchior F. (2002) Ubiquitin-related modifier SUMO1 and nucleocytoplasmic transport. Traffic 3, 381–387 [DOI] [PubMed] [Google Scholar]
- 5. Hardeland U., Steinacher R., Jiricny J., Schär P. (2002) Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 21, 1456–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Geiss-Friedlander R., Melchior F. (2007) Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell. Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 7. Dasso M. (2008) Emerging roles of the SUMO pathway in mitosis. Cell Div. 3, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Watts F. Z. (2007) The role of SUMO in chromosome segregation. Chromosoma 116, 15–20 [DOI] [PubMed] [Google Scholar]
- 9. Azuma Y., Arnaoutov A., Anan T., Dasso M. (2005) PIASy mediates SUMO-2 conjugation of Topoisomerase-II on mitotic chromosomes. EMBO J. 24, 2172–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cubeñas-Potts C., Goeres J. D., Matunis M. J. (2013) SENP1 and SENP2 affect spatial and temporal control of sumoylation in mitosis. Mol. Biol. Cell 24, 3483–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Azuma Y., Arnaoutov A., Dasso M. (2003) SUMO-2/3 regulates topoisomerase II in mitosis. J. Cell Biol. 163, 477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ryu H., Al-Ani G., Deckert K., Kirkpatrick D., Gygi S. P., Dasso M., Azuma Y. (2010) PIASy mediates SUMO-2/3 conjugation of poly(ADP-ribose) polymerase 1 (PARP1) on mitotic chromosomes. J. Biol. Chem. 285, 14415–14423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bachant J., Alcasabas A., Blat Y., Kleckner N., Elledge S. J. (2002) The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. cell 9, 1169–1182 [DOI] [PubMed] [Google Scholar]
- 14. Ryu H., Furuta M., Kirkpatrick D., Gygi S. P., Azuma Y. (2010) PIASy-dependent SUMOylation regulates DNA topoisomerase IIα activity. J. Cell Biol. 191, 783–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Porter A. C., Farr C. J. (2004) Topoisomerase II: untangling its contribution at the centromere. Chromosome Res. 12, 569–583 [DOI] [PubMed] [Google Scholar]
- 16. Hassa P. O., Hottiger M. O. (2008) The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front. Biosci. 13, 3046–3082 [DOI] [PubMed] [Google Scholar]
- 17. Song J., Durrin L. K., Wilkinson T. A., Krontiris T. G., Chen Y. (2004) Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 14373–14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kerscher O. (2007) SUMO junction-what's your function? New insights through SUMO-interacting motifs. EMBO Rep. 8, 550–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Minty A., Dumont X., Kaghad M., Caput D. (2000) Covalent modification of p73α by SUMO-1: two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J. Biol. Chem. 275, 36316–36323 [DOI] [PubMed] [Google Scholar]
- 20. Lin D. Y., Huang Y. S., Jeng J. C., Kuo H. Y., Chang C. C., Chao T. T., Ho C. C., Chen Y. C., Lin T. P., Fang H. I., Hung C. C., Suen C. S., Hwang M. J., Chang K. S., Maul G. G., Shih H. M. (2006) Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell 24, 341–354 [DOI] [PubMed] [Google Scholar]
- 21. Steinacher R., Schär P. (2005) Functionality of human thymine DNA glycosylase requires SUMO-regulated changes in protein conformation. Curr. Biol. 15, 616–623 [DOI] [PubMed] [Google Scholar]
- 22. Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S. (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141 [DOI] [PubMed] [Google Scholar]
- 23. Baumann C., Körner R., Hofmann K., Nigg E. A. (2007) PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell 128, 101–114 [DOI] [PubMed] [Google Scholar]
- 24. Kurasawa Y., Yu-Lee L. Y. (2010) PICH and cotargeted Plk1 coordinately maintain prometaphase chromosome arm architecture. Mol. Biol. Cell 21, 1188–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Leng M., Bessuso D., Jung S. Y., Wang Y., Qin J. (2008) Targeting Plk1 to chromosome arms and regulating chromosome compaction by the PICH ATPase. Cell Cycle 7, 1480–1489 [DOI] [PubMed] [Google Scholar]
- 26. Kelly A. E., Ghenoiu C., Xue J. Z., Zierhut C., Kimura H., Funabiki H. (2010) Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science 330, 235–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kornbluth S., Yang J., Powers M. (2006) Analysis of the cell cycle using Xenopus egg extracts. Curr. Protoc. Cell Biol. Chapter 11, Unit 11.11 10.1002/0471143030.cb1111s29 [DOI] [PubMed] [Google Scholar]
- 28. Ryu H., Azuma Y. (2010) Rod/Zw10 complex is required for PIASy-dependent centromeric SUMOylation. J. Biol. Chem. 285, 32576–32585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gareau J. R., Lima C. D. (2010) The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell. Biol. 11, 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ke Y., Huh J. W., Warrington R., Li B., Wu N., Leng M., Zhang J., Ball H. L., Li B., Yu H. (2011) PICH and BLM limit histone association with anaphase centromeric DNA threads and promote their resolution. EMBO J. 30, 3309–3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chan K. L., North P. S., Hickson I. D. (2007) BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 26, 3397–3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ying S., Hickson I. D. (2011) Fanconi anaemia proteins are associated with sister chromatid bridging in mitosis. Int. J. Hematol. 93, 440–445 [DOI] [PubMed] [Google Scholar]
- 33. Kaulich M., Cubizolles F., Nigg E. A. (2012) On the regulation, function, and localization of the DNA-dependent ATPase PICH. Chromosoma 121, 395–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sampson D. A., Wang M., Matunis M. J. (2001) The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 276, 21664–21669 [DOI] [PubMed] [Google Scholar]
- 35. Eladad S., Ye T. Z., Hu P., Leversha M., Beresten S., Matunis M. J., Ellis N. A. (2005) Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum. Mol. Genet. 14, 1351–1365 [DOI] [PubMed] [Google Scholar]
- 36. Wang L. H., Schwarzbraun T., Speicher M. R., Nigg E. A. (2008) Persistence of DNA threads in human anaphase cells suggests late completion of sister chromatid decatenation. Chromosoma 117, 123–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Agostinho M., Santos V., Ferreira F., Costa R., Cardoso J., Pinheiro I., Rino J., Jaffray E., Hay R. T., Ferreira J. (2008) Conjugation of human topoisomerase 2 α with small ubiquitin-like modifiers 2/3 in response to topoisomerase inhibitors: cell cycle stage and chromosome domain specificity. Cancer Res. 68, 2409–2418 [DOI] [PubMed] [Google Scholar]
- 38. Biebricher A., Hirano S., Enzlin J. H., Wiechens N., Streicher W. W., Huttner D., Wang L. H., Nigg E. A., Owen-Hughes T., Liu Y., Peterman E., Wuite G. J., Hickson I. D. (2013) PICH: a DNA translocase specially adapted for processing anaphase bridge DNA. Mol. Cell 51, 691–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nathan D., Ingvarsdottir K., Sterner D. E., Bylebyl G. R., Dokmanovic M., Dorsey J. A., Whelan K. A., Krsmanovic M., Lane W. S., Meluh P. B., Johnson E. S., Berger S. L. (2006) Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev. 20, 966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]