FIGURE 2.
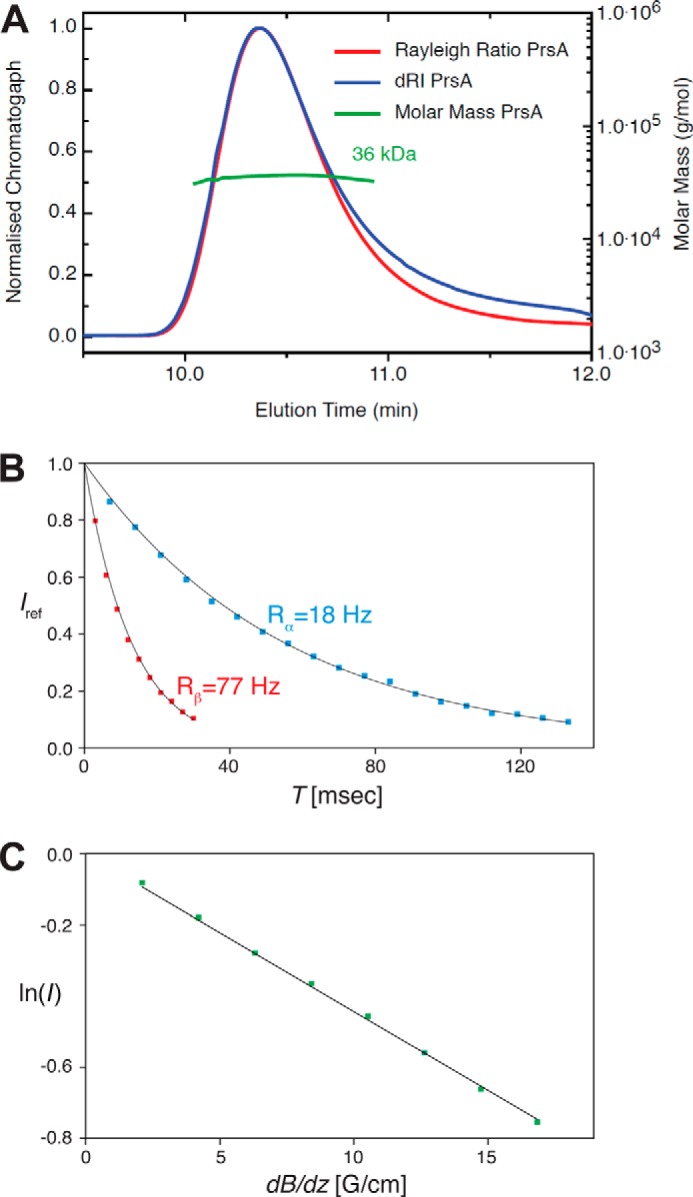
Molecular weight and biophysical molecular parameter determination of PrsA. A, size exclusion chromatography coupled to SEC-MALS of PrsA. The molecular mass was calculated throughout the eluting peaks and is indicated in green. The calculated mass of 36.0 kDa is in good agreement with the 33.4 kDa for the N-terminal His and tobacco etch virus protease cleavage site-tagged (MHHHHHHSSGVDLGTENLYFQ*SM (*, tobacco etch virus protease cleavage site) PrsA protein (residues 21–292). B, [15N,1H]-TRACT experiment (80) for the determination of the effective rotational correlation time constant τc. The one-dimensional proton signal intensity, Iref, of a sample of 750 μm PrsA at 25 °C was integrated between 10 and 8.5 ppm and plotted versus the relaxation period T. The transverse relaxation rates of the TROSY component (blue; black: least squares fit) and the anti-TROSY component (red; black: least squares fit) were determined to be Rα = 18 Hz and Rβ = 77 Hz, resulting in τc = 23 ns, that is typical for a globular protein of about 60 kDa size (81). C, measurement of the molecular diffusion constant in aqueous solution by the 15N-filtered diffusion BPP-LED NMR experiment (82) for 750 μm PrsA (green). The logarithm of the signal intensity, integrated between 10 and 8.5 ppm, is plotted versus the gradient strength of the pulsed field gradients. The black line represents the linear fit to the measured data. This measurement yielded the self-diffusion constants, D0 = 6.11 × 10−11 m2 s−1, that corresponds under the assumption of a spherical molecule and by using the Stokes-Einstein equation to an estimated molecular mass of 58 kDa.
