FIGURE 6.
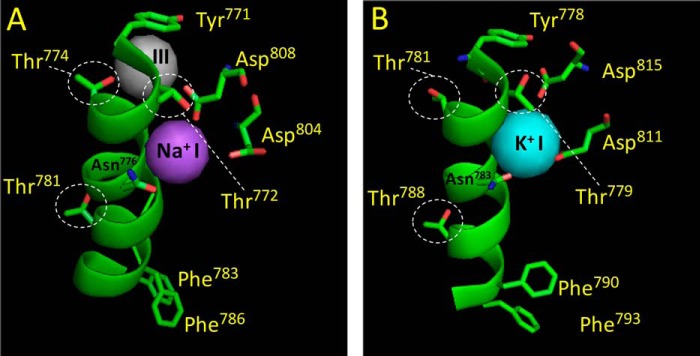
Rotamer transitions revealed by the high-resolution crystal structures of the Na+,K+-ATPase. Site I coordination in the E1·AlF4−·ADP·3Na+ structure (A, PDB code 3WGU, Ref. 18), and the E2·MgF42−·2K+ structure (B, PDB code 2ZXE, Ref. 30). The figure shows site I and the position of important M5 residues as well as the side chains of Thr781, Phe783, and Phe786 (kidney sequence), located near the extracellular surface. A, the hydroxyl group of Thr772 is coupled to a Na+ in site I. The hydroxyl group of Thr774 is contacting a Na+ in site III (shown in gray). The side chains of Phe783 and Phe786 bend toward the extracellular side. B, the methyl group of Thr772 is connected to a K+ in site I through water molecules. The side chain of Thr781 is disconnected from “empty site III.” The side chains of Phe790 and Phe793 (shark sequence) are moved toward the cytoplasm (compare with their position to that in the E1 structure). Note that the side chain of Thr781 (Thr788 in the shark sequence), located away from the cytoplasmic side, does not adopt rotamer transition, i.e. the side chain has the identical position in both structures. Note the close proximity of Asp808 (Asp815 in the shark sequence) to the Na+ in site I compared with its position in the E2 structure, this insertion (likely facilitated by the movement of M6) seems to regulate the traffic of water molecules into the binding cavity around site I. Note also that the side chain of Asn776 also seems to adopt a rotation, directing the side chain hydroxyl toward K+ in site I in the E2 structure (Asn783 in shark sequence). The figure was made using PyMOL.
