Abstract
Growing evidence indicates that RNA oxidation is correlated with a number of age-related neurodegen-erative diseases, and RNA oxidation has also been shown to induce dysfunction in protein synthesis. Here we study in vitro RNA oxidation catalyzed by cytochrome c (cyt c)/H2O2 or by the Fe(II)/ascorbate/H2O2 system. Our results reveal that the products of RNA oxidation vary with the oxidant used. Guanosine residues are preferentially oxidized by cyt c/H2O2 relative to the Fe(II)/ascorbate/H2O2 system. GC/MS and LC/MS analyses demonstrated that the guanine base was not only oxidized but also depurinated to form an abasic sugar moiety. Results from gel electrophoresis and HPLC analyses show that RNA formed a cross-linked complex with cyt c in an H2O2 concentration-dependent manner. Furthermore, when cyt c was associated with liposomes composed of cardiolipin/phosphatidylcholine, and incubated with RNA and H2O2, it was found cross-linked with the oxidized RNA and dissociated from the liposome. Results of the quantitative analysis indicate that the release of the cyt c from the liposome is facilitated by the formation of an RNA–cyt c cross-linked complex. Thus, RNA oxidation may facilitate the release of cyt c from the mitochondrial membrane to induce apoptosis in response to oxidative stress.
Keywords: RNA oxidation, Cytochrome c release, Cross-link, Abasic site, 8-Hydroxyguanosine, Free radicals
Introduction
Free radicals and reactive oxygen species (ROS)2 play major roles in normal biological functions. However, they also react rapidly with biomacromolecules such as nucleic acids, lipids, and proteins, which leads to a loss of or change in their biological functions. Thus, they have been implicated in a wide range of age-related neurodegenerative conditions and in aging (for reviews see [1-3]). Among nucleic acid oxidations, much attention has been focused on DNA, which is protected by its binding proteins and by a number of repair systems to minimize damaging effects [4]. Because RNAs are less protected against ROS, they are more susceptible to oxidation than DNA [5]. Growing evidence indicates that age-related diseases, including atherosclerosis [6], dementia with Lewy bodies [7], Parkinson disease [8], amyotrophic lateral sclerosis [9], and Alzheimer disease [10], are correlated with elevated levels of oxidized RNA. Furthermore, it has been reported that during the very early stages of Alzheimer disease, protein synthesis declines, whereas RNA oxidation increases, suggesting that oxidized RNA could be one of the primary causes for the disease pathogenesis [11-13]. Despite the fact that there is sufficient evidence implicating RNA oxidation with various age-related neurodegenerative diseases and other ROS-induced physiological damage, knowledge of the nature of RNA oxidation is quite limited, particularly compared to that reported for DNA oxidation. In fact most RNA oxidation studies are based on monitoring the formation of 8-hydroxyguanosine (8-OH-Guo).
Studies of DNA oxidation reveal that in addition to the oxidation of bases, depurination and depyrimidination are major oxidative modifications. To detect the abasic deoxyribose moiety after depurination/depyrimidination, Ide et al. [14] developed an aldehyde-reactive probe (ARP) using N′-aminooxymethylcarbo-nylhydrazino d-biotin to react with the aldehyde moiety in the abasic sugar derivatives. We found that ARP reacts quantitatively to as little as 10 fmol of depurinated tRNAPhe or chemically synthesized RNA containing abasic sites, indicating that abasic RNA is recognized by ARP. In vitro experiments showed that ARP reacted effectively with RNA oxidized by Fenton-type reactions, γ-irradiation, or peroxynitrite. Furthermore, ARP reactivity was also found to be elevated when it reacted with cellular RNA obtained from cultured cells that had been subjected to oxidative stress conditions. Using ARP we have developed a method to isolate and quantify oxidized levels of specific RNA species [15]. Together, these observations indicate that ARP can serve as a sensitive probe for RNA oxidation induced by ROS or reactive nitrogen species because abasic residues are generated during RNA oxidation [16].
In vitro DNA oxidation mediated by metal ions or carcinogenic agents has been shown not only to generate discrete oxidized nucleotides but also to promote DNA cross-linking with proteins [17-19]. In addition, formation of DNA–protein cross-links between DNA polymerase β, the repair enzyme that catalyzes the excision of abasic sites, and the C1′-oxidized abasic residue (2-deoxyribonolac-tone; dL) of oxidized DNA has been reported [20]. These authors showed that dL functions as a suicide substrate by forming an amide linkage with the lysyl-72 residue of the polymerase and inhibiting its activity. In the case of protein–RNA cross-linking, Mirzaei and Regnier [21] reported that several ribosomal proteins and rRNA were cross-linked when yeasts were treated with H2O2, suggesting that cross-linking between RNA and protein occurs under oxidative stress conditions.
Cytochrome c (cyt c) is known to possess peroxidase activity. This positively charged cyt c readily associates with cardiolipin, the acidic lipid in the mitochondrial inner membrane [22], and the negatively charged nucleic acid [23], as shown in the case of tRNA [24], through electrostatic interaction. Thus, in the presence of H2O2 these molecules are preferentially oxidized by the cyt c-mediated catalytic pathway. In fact, cyt c has been shown to induce peroxidation of cardiolipin [25] and ROS have also been implicated in the dissociation of cyt c from the inner mitochon-drial membrane[26], suggesting that peroxidation of cardiolipin catalyzed by cyt c may play an important role in cyt c release into the cytoplasm during the apoptotic process. However, there has been no investigation into the physiological effects of cyt c-mediated RNA oxidation.
In this study, we report that RNA oxidized by cyt c-mediated peroxidation (compared to the Fenton reaction) leads to depurination of guanosine and abasic formation. However, 8-OH-Guo is not detected when RNA is oxidized by the H2O2/cyt c system. Furthermore, under these experimental conditions, cyt c is found cross-linked with RNA in an H2O2 concentration-dependent manner. This RNA-cross-linked cyt c appears to facilitate the dissociation of cyt c from the liposome that is composed of cardiolipin/phosphatidyl-choline. Based on our findings, the deficiency of monitoring RNA oxidation based solely on 8-OH-Guo measurement and the potential role of the RNA–cyt c cross-linked complex generated during cyt c-catalyzed peroxidation of RNA in the cyt c release from the mitochondrial membrane to initiate apoptosis under oxidative stress conditions are discussed.
Materials and methods
Reagents
All the buffer solutions for the RNA synthesis, isolation, and oxidation were pretreated with Chelex 100 (Bio-Rad) or contained 1 mM EDTA as indicated. All the reagents used were purchased from Sigma–Aldrich unless otherwise specified.
Preparation of RNA
The RNA synthesis reaction was initiated by incubating a DNA template encoding a FLAG-tagged luciferase 2 gene (approximately 1.9 kb) with T7 RNA polymerase (MEGAscript kit; Ambion, Austin, TX, USA). For radiolabeled RNA, 5 μCi of [8′-14C]guanosine triphosphate (Moravek Biochemicals, Brea, CA, USA) was added to the reaction mixture including the DNA template encoding part of human 18 S rRNA (128 bases) included in the MEGAshortscript kit (Ambion). After incubation for up to 4 h at 37 °C, followed by incubation with DNase for 10 min, the synthesized RNA was purified with the PureLink total RNA isolation kit (Invitrogen, Carlsbad, CA, USA) and eluted with RNase-free water according to the manufacturer’s protocol. The synthesized RNA (250 μg/ml) was reacted with either H2O2 and 1/1 Fe(II)/ascorbate mixture or H2O2 and bovine cyt c in 10 mM Hepes (pH 7.4) buffer for 1 h at 37 °C. The oxidation reaction was terminated by adding 5 mM EDTA, then quickly precipitated with 2.5 vol of cold ethanol and 0.1 vol of 3 M sodium acetate, and stored at −80 °C until use. Precipitated RNA was suspended in 10 mM Tris–HCl (pH 8.0), 1 mM EDTA (TE buffer) for analysis.
High-performance liquid chromatography (HPLC) analysis of hydrolyzed RNA
HPLC analyses were performed on an Agilent Model 1100 system (Agilent Technology, Santa Clara, CA, USA) equipped with an Agilent G1315A diode array detector and G1311A quaternary pump equipped with Agilent ChemStation software. Based on a previous report [27], hydrolysis of oxidized RNA was performed as follows: 20 μg of RNA suspended in 10 mM ammonium acetate (pH 5.3) was digested with 1 U of P1 nuclease at 37 °C for 1 h. One-tenth volume of 1 M freshly prepared ammonium bicarbonate and 0.1 U of phosphodiesterase were added to the reaction mixture and incubated at 37 °C for 1 h, followed by treatment with 2 U of alkaline phosphatase at 37 °C for 1 h. An aliquot of the hydrolysate was applied to a preheated (30 °C) RP-18 HPLC column (4.6 × 250 mm, 5.0-μm particle size) (Waters Corp., Milford, MA, USA) equilibrated with buffer A, 0.1% trifluoroacetic acid (TFA), and eluted at 1 ml/min with buffer A and then with buffer B, 80% acetonitrile, 0.1% TFA, with a gradient shown in Fig. 5. Gradient conditions were as follows: 0–8 min, linear gradient from 0 to 2% buffer B; 8–24 min, gradient from 2 to 12.5% buffer B; 24–30 min, gradient from 12.5 to 80%; 30–45 min, hold at 80% buffer B. Each ribonucleoside was quantified with the diode array detector to monitor absorbance at 260 nm. To examine oxidation of guanosine, radiolabeled RNA with [8′-14C]guanosine was enzymatically hydrolyzed and applied to the HPLC. Fractions were collected every 30 s with a FoxyJr (Isco, Lincoln, NE, USA) fraction collector, and the radioactivity of each fraction was measured using a scintillation counter (Beckman, Brea, CA, USA). UV absorbance spectrum from 200 to 400 nm was monitored as necessary.
Fig. 5.
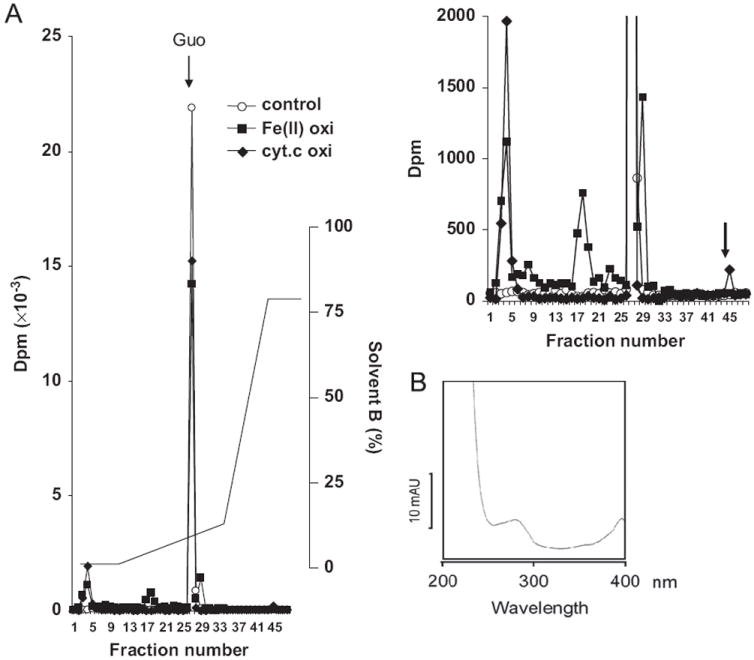
Tracer experiment using RNA labeled with [14C]guanine. RNA radiolabeled with [14C]guanosine was oxidized by either 20 μM/1 mM of cyt c/H2O2 or 20 μM/20 μM/1 mM of Fe(II)/ascorbate/H2O2. After hydrolysis, oxidized guanosine was separated using an RP-18 column. (A) Radioactivity measured in each fraction and the HPLC gradient pattern. A chromatogram with expended scale of the y axis is shown at the upper right. (B) The radioactive fraction (No. 45) indicated by the arrow in the RNA oxidized by cyt c shows a spectrum similar to that of ferricytochrome c.
Quantification of oxidized base derivatives by GC/MS
To quantify 8-OH-Guo, 20 μg of oxidized RNA was lyophilized overnight together with stable isotope internal standards including 0.2 nmol of 8-hydroxydeoxyguanosine (8-OH-dGuo)-13C2,15N2 and 20 nmol of deoxyguanosine-15N5. The RNA was hydrolyzed with 0.5 ml of 60% formic acid in evacuated, sealed tubes at 140 °C for 30 min and then lyophilized. The hydrolysate was trimethylsilylated and then applied to GC/MS according to the procedures reported previously [28]. For quantification of 8-hydroxyguanine (8-OH-Gua) cleaved from the RNA chain, oxidized RNA was precipitated with ethanol. The supernatant fraction was mixed with 0.1 nmol of internal standard of 8-OH-Gua-13C2,15N2 and then lyophilized without hydrolysis. The mixture was trimethylsilylated and then applied to GC/MS.
Identification of abasic sites by LC/MS
Twenty micrograms of RNA was oxidized by 100 μM Fe(II)/ascorbate and H2O2 or cyt c–H2O2 as described above. Synthetic oligo RNA containing abasic sites was prepared using the method reported previously [29]. Oxidized RNA or oligo RNA was derivatized with ARP as described previously [16] and then mixed with formaldehyde to terminate the reaction. After RNA was enzymatically hydrolyzed [27], 0.4 μg of hydrolysate was applied to a C-18 capillary column (Magic MS C18; 5-μm particle size, 200-Å pore size, 0.5 × 120 mm) and eluted with solution A (0.09% formic acid, 0.01% TFA, 2% acetonitrile) and solution B (0.09% formic acid, 0.0085% TFA, 95% acetonitrile) with the following gradients: 0–10 min, linear gradient from 2 to 35% solution B; 10–12 min, gradient from 35 to 60% solution B; 12–25 min, hold at 100% solution B, with a flow rate of 20 μl/min. Mass spectrometric analyses were performed with the positive ion mode using an HCT Ultra Ion trap system (Bruker Daltonics, Billerica, MA, USA). The nozzle-skimmer voltage was set at 40 V, the capillary voltage at 4000 V, and desolvation temperature at 300 °C, and the flow rate was 10 L/min.
Cross-linking between RNA and cyt c
Short RNA (128 bases) was synthesized in the presence of 50 μCi of [32P]CTP (PerkinElmer, Waltham, MA, USA) as described above. The radiolabeled RNA was oxidized with cyt c and H2O2 at 37 °C for 1 h. The reaction mixture was transferred into Laemmli sample buffer and heated at 95 °C for 5 min and then applied onto a 4–12% gradient SDS–PAGE gel. After electrophoresis the gel was fixed with 10% methanol and 10% acetic acid for 20 min, followed by staining with Coomassie brilliant blue (CBB). The gel was then dried overnight on a filter paper using a gel dryer. The dried gel was exposed on a phosphorimager screen (GE Healthcare).
Dissociation of cyt c from liposomes
Based on a previous report [30], cardiolipin (CL) derived from bovine heart (Avanti, Alabaster, AL, USA) or phosphatidylcholine (PC) derived from egg yolk was suspended in a chloroform/methanol mixture at a concentration of 20 mg/ml. CL (25%) and PC (75%) were mixed in a glass tube and evaporated by flashing nitrogen gas for 30 min to make a thin film and then subjected to vacuum evaporation for 4 h. The lipid film was suspended in Hepes buffer containing 10 mM Hepes (pH 7.4), 1 mM EDTA, to a final concentration of 10 mg/ml total lipid. The mixture was vortexed occasionally for 1 h. The multilamellar liposomes were centrifuged at 2000g for 5 min to remove clogged lipid, then precipitated by ultracentrifugation at 100,000g for 15 min, and resuspended in Hepes buffer and stored at − 10 °C until use.
The liposomes (400 μg) were mixed with bovine cyt c (1 nmol) for 10 min at room temperature, and the free cyt c was removed by ultracentrifugation (this process was repeated three times) as described above. The cyt c-containing liposomes were suspended in 25 μl of Hepes buffer containing 10 mM Hepes (pH 7.4) and 1 mM deferoxamine. Forty or 400 μg/ml yeast tRNA and 50, 200, or 800 μM H2O2 as indicated were mixed simultaneously with cyt c-containing liposomes and incubated at 37 °C for 40 min. The reaction mixtures were then subjected to ultracentrifugation to separate the precipitated liposome from the supernatant fraction. The precipitant fraction was resuspended in 25 μl of pH 7.4 Hepes buffer. Eight microliters from each fraction was mixed with Laemmli sample buffer at 95 °C for 5 min and loaded onto an SDS–polyacrylamide gel. After electrophoresis, the gel was stained with Imperial CBB staining solution (Invitrogen) according to the manufacturer’s recommended protocol to monitor the cyt c. The levels of cyt c and cyt c-containing bands were monitored using an Odyssey (Li-Cor) fluorescence detector at 700 nm wavelength window. Densitometric method was used to quantitate the intensity observed for species > 10 kDa in each fraction using the software provided by Li-Cor. The dissociated cyt c was expressed as the ratio of intensity detected in the supernatant fraction to that of the precipitant fraction.
Statistical analyses
The experimental results were statistically analyzed using one-way ANOVA and Sigma Stat software.
Results
Oxidative modification of RNA by Fe(II) or cytochrome c
Because cyt c possesses peroxidase activity and it also readily interacts with negatively charged nucleic acids, we carried out a comparative study of RNA oxidation catalyzed by Fe(II)/ascorbate and by cyt c. In this study, an in vitro synthesized RNA was oxidized with either cyt c or Fe(II)/ascorbate in the presence of H2O2 at 37 °C for 1 h. The resulting oxidized RNA was hydrolyzed to monoribonucleosides by P1 nuclease, phosphodiesterase, and alkaline phosphatase; each ribonucleoside was separated with reverse-phase HPLC. The results shown in Fig. 1A indicate that a number of ribonucleoside peaks obtained from the oxidized RNA hydrolysate were decreased compared to those observed with nonoxidized RNA hydrolysate. In addition, some minor peaks were also observed in the sample when RNA was oxidized by the Fe(II)/ascorbate/H2O2 system (Fig. 1A). A relative decrease in ribonucleosides caused by either Fe(II)/ascorbate or cyt c treatment is shown in Fig. 1B, as a function of increasing H2O2 concentration. Overall the data reveal that all ribonucleosides were reduced readily by H2O2 when Fe(II)/ascorbate was used as a catalyst. However, with cyt c as catalyst, guanosine is by far the predominant ribonucleoside oxidized (Fig. 1C).
Fig. 1.
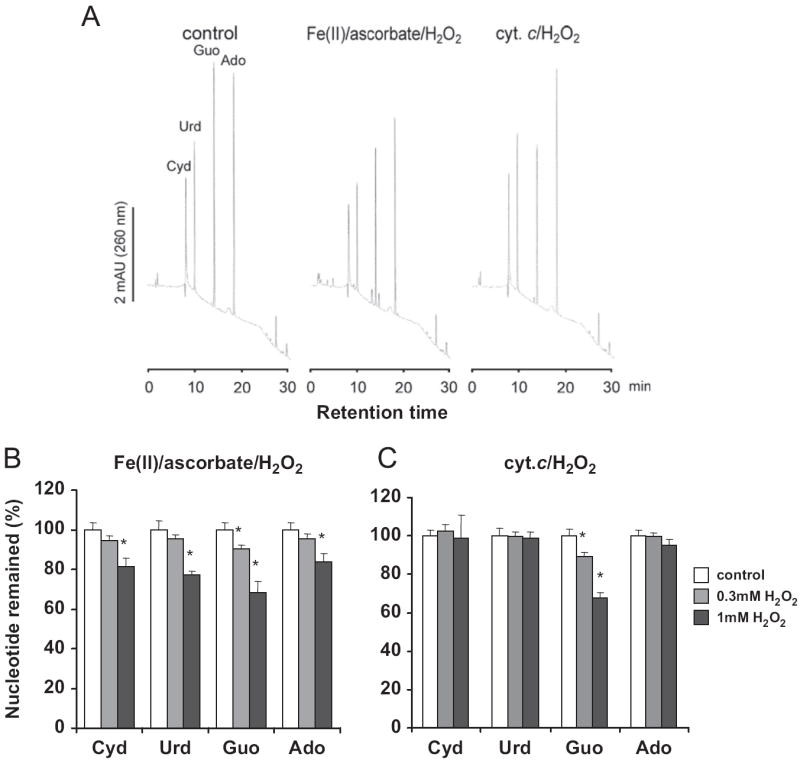
Oxidative modification of ribonucleosides by Fe(II) or cyt c. (A) In vitro synthesized RNA was oxidized with 20 μM cyt c or 20 μM iron/ascorbate in the presence of 1 mM H2O2 for 1 h at 37 °C. The enzymatically hydrolyzed RNA was applied to a reverse-phase HPLC column and absorbance monitored at 260 nm. Recovery of each ribonucleoside after oxidation with the indicated concentration of H2O2(no added H2O2 for control), catalyzed (B) by 6 μM/6 μM of Fe(II)/ascorbate, or (C) by 6 μM of cyt c was quantified by the peak volume. The values shown represent means ± SD from triplicate samples. *p < 0.05 relative to the nonoxidized control. Ado, adenosine; Cyd, cytidine; Guo, guanosine; Urd, uridine.
Quantitation of 8-OH-Guo in RNA
8-OH-Guo has been regarded as a major oxidized derivative in oxidized RNA. To quantify the formation of 8-OH-Guo as a form of ribonucleotide in the oxidized RNA catalyzed either by Fe(II)/ascorbate or by cyt c, the oxidatively modified RNA was analyzed with the stable isotope dilution method using a GC/MS machine as described earlier [28]. The results shown in Fig. 2 indicate that 8-OH-Guo is generated as a function of increasing H2O2 concentration when RNA oxidation is catalyzed by Fe(II)/ascorbate, whose activity is greatly inhibited by the addition of 5 mM EDTA. In contrast, there was no significant increase in 8-OH-Guo detected in the cyt c-catalyzed oxidation of RNA, even when the concentration of cyt c and H2O2 was elevated to 20 μM and 1 mM, respectively.
Fig. 2.
Quantification of 8-OH-Guo in oxidized RNA. In vitro synthesized RNA was oxidized using the indicated concentrations of Fe(II)/ascorbate/H2O2 or cyt c/H2O2 for 1 h at 37 °C. The oxidized RNA was monitored using the GC/MS method together with the internal standard as described under Materials and methods. The results represent means ± SD in triplicate experiments. *p < 0.05 relative to the nonoxidized control.
8-OH-Guo was depurinated after oxidation
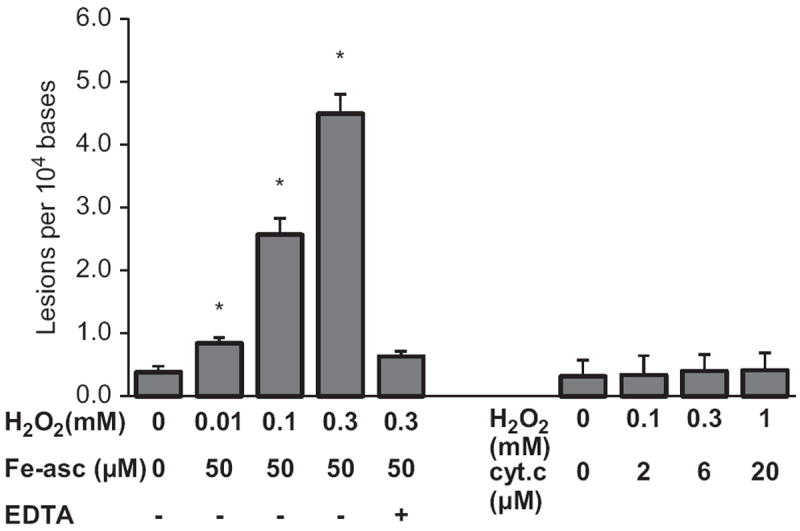
Fig. 1 shows that guanosine is readily oxidized by either Fe(II)/ascorbate or cyt c catalysis, yet no significant quantity of 8-OH-Guo was detected in the RNA oxidized by the cyt c/H2O2 system (see Fig. 2). Thus, we examined whether 8-OH-Guo was further depurinated after oxidation by either cyt c/H2O2 or Fe(II)/ascorbate/H2O2. To this end, oxidized RNA was precipitated with ethanol and the supernatant fraction was analyzed using a GC/MS machine, together with the internal 8-OH-Gua standard without acid hydrolysis. The results shown in Fig. 3 reveal that 8-OH-Gua was detected in the supernatant fraction of the oxidized RNA catalyzed by either Fe(II)/ascorbate or cyt c, and its quantity is H2O2 concentration dependent. Furthermore, the levels 8-OH-Gua generated are significantly higher in the cyt c/H2O2 oxidizing system relative to those obtained in the Fe(II)/ascorbate/H2O2 system. It should be pointed out that the quantity of 8-OH-Gua detected in the supernatant fraction does not contain the contribution from 8-OH-Guo in RNA, because acid hydrolysis was omitted in the sample preparation. Together, these results reveal that guanine base was not only oxidized to 8-OH-Gua but also cleaved from the RNA during its oxidation catalyzed by either Fe(II)/ascorbate or cyt c.
Fig. 3.
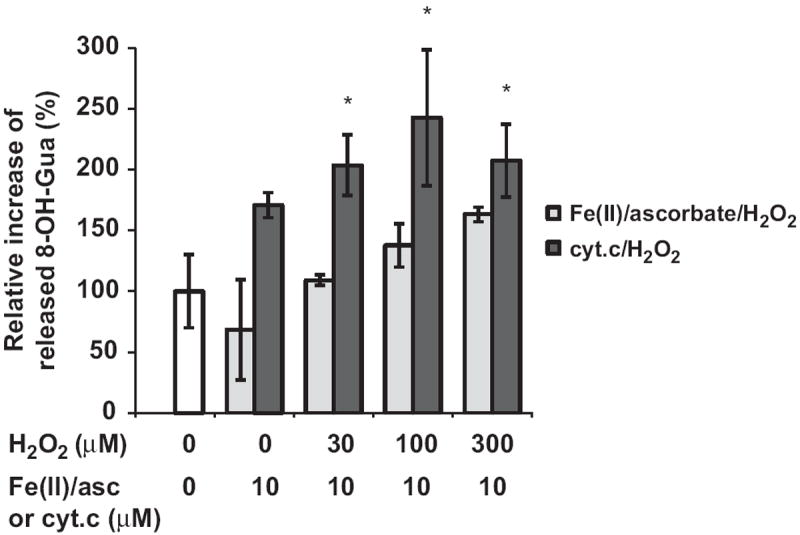
Quantification of cleaved 8-OH-Gua. After in vitro oxidation with 10 μM Fe(II)/ascorbate or 10 μM cyt c and the indicated concentrations of H2O2, oxidized RNA was precipitated with ethanol. The supernatant was mixed with internal standards and then trimethylsilylated without acid hydrolysis. The resulting mixture was analyzed with GC/MS. The quantities of 8-OH-Gua generated are presented as percentage increase relative to that observed with control. The results represent means ± SD in triplicate experiments. *p < 0.05 relative to the nonoxidized control.
Generation of abasic sites during cyt c-catalyzed RNA oxidation
Because the cyt c-catalyzed RNA oxidation leads to depurination of 8-OH-Guo, it is assumed that under these conditions abasic sites are also generated. To verify this notion, oxidized RNA was derivatized with an ARP, which reacts with the aldehyde moiety in abasic sites [14,16]. The product of the ARP-derivatized abasic site, which is equivalent to an ARP–ribose conjugate, was detected in RNA oxidized by cyt c. The extracted ion chromatograms at 464.2 ± 0.3 m/z corresponding to ARP-abasic are shown in Fig. 4A. The results from LC/MS analysis are shown in Fig. 4B and reveal that the ARP-abasic derivative generated by cyt c-mediated oxidation of RNA and those formed with Fe (II)/ascorbate/H2O2 and positive control, derived from synthetic abasic RNA, all exhibit a retention time of 8.5 min. These data demonstrate that RNA abasic sites were generated by cyt c-catalyzed RNA peroxidation.
Fig. 4.
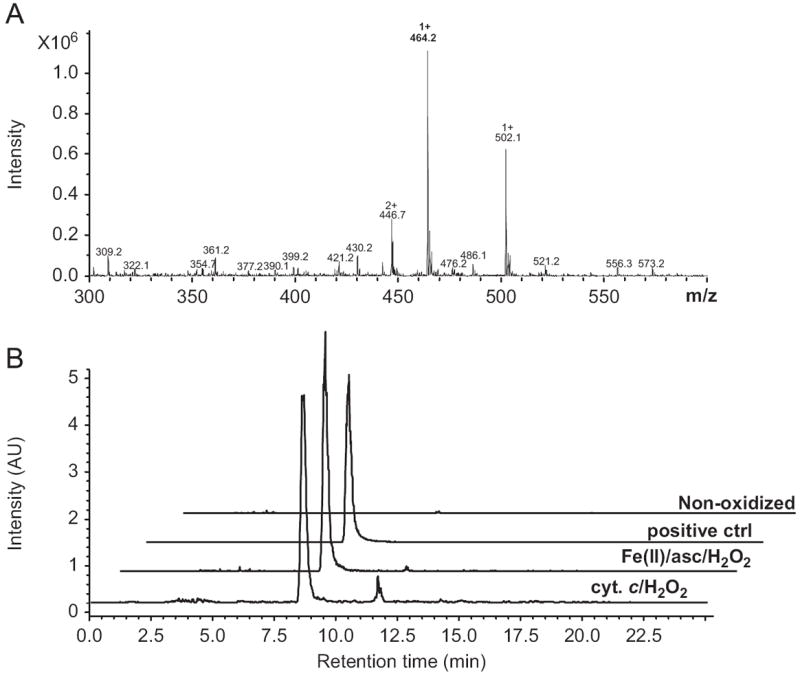
Identification of abasic site produced by cyt c. RNA oxidized by cyt c was derivatized with ARP and hydrolyzed. The hydrolysate was applied to LC/MS to identify ARP-derivatized abasic sites. As a reference, nonoxidized RNA with or without synthetic abasic sites or RNA oxidized by Fe(II)/ascorbate/H2O2 was eluted and analyzed under the same conditions. (A) The extracted ion chromatograms at 464.2 ± 0.3 m/z corresponding to ARP-abasic. (B) The mass spectra of the oxidized RNA in the presence of 20 μM/1 mM cyt c/H2O2 for 1 h at 37 °C eluted at 8.5 min.
Cross-link between RNA and cyt c
In view of the fact that cyt c-mediated RNA oxidation generates abasic sites and the aldehyde moieties on the abasic sites can readily form a Schiff base with the amino groups in proteins, we investigated the possibility that oxidized RNA could cross-link with cyt c. To this end RNA radiolabeled with [8′-14C]guanosine was oxidized by either cyt c/H2O2 or Fe(II)/ascorbate/H2O2. After enzymatic hydrolysis, the oxidized guanosine residue was separated by HPLC similar to the methods used to obtain the data given in Fig. 1. As shown in Fig. 5A, the radiolabeled products generated with Fe(II)/ascorbate/H2O2 oxidation yielded several low-level radioactive species eluted from fraction 5 through fraction 25, with additional radioactive peaks in fractions 3, 17, and 28. The elution pattern of these radioactive peaks is consistent with the HPLC result shown in Fig. 1A, indicating that several oxidized derivatives were generated by iron/ascorbate-mediated oxidation. However, there are only two radiolabeled fractions generated by cyt c-mediated oxidation. One of these fractions was eluted in the void fraction and the other, fraction 45, eluted with a high concentration of acetonitrile (see Fig. 5A and its expanded y-scale chromatogram shown in the upper right). The total amount of radioactivity eluted after cyt c-mediated peroxidation was approximately 20% lower than that obtained with control, whereas in the case of Fe(II)/ascorbate-mediated oxidation the total eluted radioactivity was reduced only moderately (approximately 6% reduction). Interestingly, the peak in fraction 45 was found to contain cyt c, because its spectrum exhibits a characteristic absorbance spectrum of ferricytochrome c with an absorbent peak at 395 nm (Fig. 5B). It should be pointed out that the absorption spectrum was obtained with sample eluted with solvent containing 0.1% TFA, which has a pH of 2.0. Under this acidic condition, the Soret peak of cyt c occurs at 395 nm [31]. Thus, these observations indicate that the radiolabeled guanosine and cyt c form a covalently linked complex.
To confirm that cyt c-mediated peroxidation leads to RNA cross-linking with cyt c, 32P-radiolabeled RNA and cyt c were incubated for 1 h at 37 °C with H2O2 at the indicated concentrations. The reaction mixture was applied to SDS–PAGE, followed by autoradiography to identify RNA-containing fractions (Fig. 6A) and CBB staining to identify cyt c protein-containing fractions (Fig. 6B). These results show that in the presence of RNA and H2O2, RNA and cyt c comigrated as a high-molecular-weight species (lanes 3 and 4), indicating that RNA and cyt c were covalently bonded. In the absence of RNA, cyt c did not form a high-molecular-weight complex; it eluted as monomeric cyt c. Interestingly, the quantity of this monomeric cyt c decreased as a function of increasing H2O2 concentration, probably caused by its oxidative degradation (lanes 5, 6, and 7). Together, these results indicate that RNA and cyt c indeed form a cross-linked complex in the presence of H2O2.
Fig. 6.
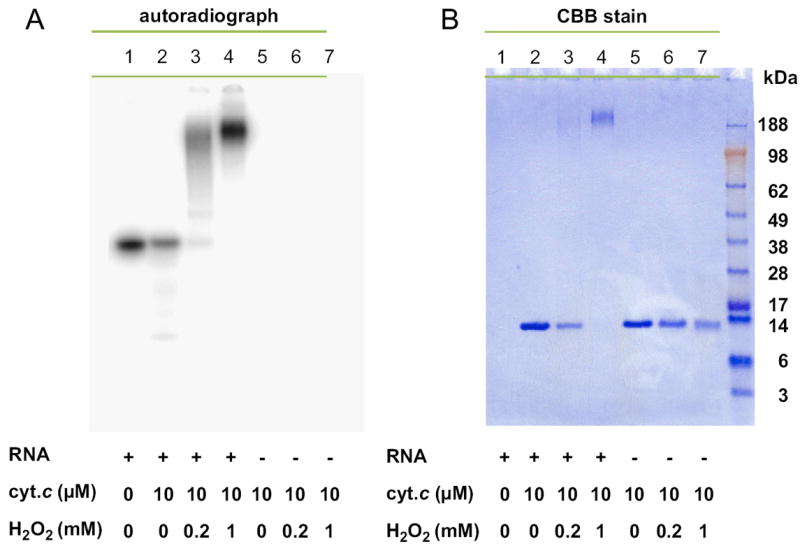
Cross-linking between RNA and cyt c. Radiolabeled RNA and/or cyt c was incubated with H2O2 at the indicated concentrations for 1 h at 37 °C. The reaction mixture was heat denatured and loaded onto an SDS–PAGE gel. The gel was (A) autoradiographed or (B) stained with CBB.
Release of cyt c from liposomes is mediated by oxidative interaction with RNA
To investigate the potential physiological function of cross-linked RNA–cyt c, we examined the oxidative interaction between RNA and cyt c bound to the liposome membrane containing CL. Cyt c was associated with a multilamellar liposome composed of 25% CL and 75% PC. This association of cyt c was specific for CL, because cyt c did not associate with liposomes composed of only PC (data not shown). The cyt c-loaded liposomes were incubated with tRNA and H2O2 for 40 min at 37 °C, followed by ultracentrifugation to separate the supernatant and liposomes. Each fraction was loaded onto an SDS–PAGE gel, followed by staining with CBB to detect cyt c-containing complexes. Fig. 7A depicts the CBB-detected cyt c and cyt c-containing complexes that migrated to the supernatant fraction during the H2O2-mediated oxidation in the presence or absence of tRNA as indicated. Note that we attributed all protein-containing bands to a cyt c origin because the only protein in the reaction mixture was cyt c. The cyt c and cyt c-containing complexes remaining in the liposome after oxidation are shown in Fig. 7B. Overall, the percentages of cyt c dissociated from the liposome induced by cyt c/H2O2-mediated oxidation are presented in Fig. 7C. Note that the results depicted in the first four lanes of Fig. 7B indicate that cross-link formation was detected after treatment of cyt c with H2O2, possibly because of cyt c cross-linking with CL. However, the data for these corresponding lanes shown in the first four lanes in Fig. 7C indicate that cyt c/H2O2-mediated CL oxidation alone does not induce extensive release of cyt c from liposomes. This observation is consistent with recent reports showing that although CL is oxidized by a cardiolipin-specific peroxidase of CL-bound cyt c [25], the release of cyt c in model membranes and in brain mitochondria seems to be independent of CL peroxidation by H2O2 [32]. However, in the presence of both tRNA and H2O2, the quantity of cyt c together with its cross-linked complexes observed in the supernatant fraction was significantly elevated (Fig. 7C). Thus, tRNA oxidized by cyt c/H2O2 induces the release of cyt c from CL-containing liposomes, probably mediated by cyt c cross-linking with the oxidized tRNA.
Fig. 7.
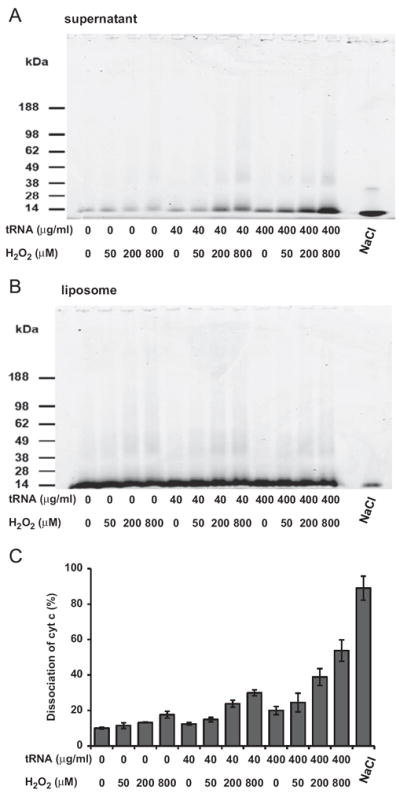
H2O2 and RNA facilitate the dissociation of cyt c from liposomes. Bovine cyt c (1 nmol) was associated with 25 μl of liposomes composed of cardiolipin (25%)/phosphatidylcholine (75%) by mixing the cyt c with the liposomes for 10 min at room temperature. The free cyt c was removed by ultracentrifugation three times as described under Materials and methods. After 40 min incubation at 37 °C with the indicated concentrations of tRNA and/or H2O2 or 0.4 M NaCl, the reaction mixtures were ultracentrifuged to separate the supernatant fraction from the liposome fraction. Both fractions were heat denatured and then loaded onto an SDS–PAGE gel. Protein bands (cyt c-containing bands) in the supernatant and in liposomes were stained with CBB and are shown in (A) and (B), respectively. (C) The intensities of cyt c and cyt c-containing bands were monitored using the Odyssey (Li-Cor) method. A densitometric method, based on the intensity observed for species > 10 kDa in both fractions, was used to quantify all the cyt c-containing species in the supernatant and in the liposomes and expressed as the ratio of cyt c dissociated from the liposomes. The values shown represent means ± SD from triplicate experiments.
Discussion
In this study, we examined RNA oxidation catalyzed either by Fenton reaction mediated by Fe(II)/ascorbate/H2O2 or by cyt c/H2O2. Our results reveal that (i) although guanosine is preferentially oxidized among the four nucleosides, the oxidized products vary with the oxidants used. As a result it is necessary to use multiple oxidation markers to assess RNA oxidation. (ii) RNA oxidation mediated by cyt c leads to the formation of cross-linking complexes between RNA and cyt c. (iii) RNA and H2O2 facilitate cyt c release from liposomes containing CL, in part as a cross-linked complex with RNA. Thus, RNA oxidation may be involved in facilitating the oxidative stress-induced cellular apoptosis.
It is generally assumed that 8-OH-dGuo and 8-OH-Gua are appropriate oxidation markers for DNA and RNA, respectively. However, recent reports indicate that 8-OH-dGuo may not be sufficient to evaluate oxidative damage of DNA, because 8-OH-dGuo has a low redox potential and is readily subjected to further oxidation [33]. Furthermore, oxidized derivatives other than 8-OH-dGuo have been reported to be predominantly generated depending on the oxidative stress conditions [34]. Similarly, in the case of RNA oxidation, whether monitoring the formation of 8-OH-Guo alone is sufficient to address its oxidative damage remains to be investigated. Our results show that the oxidized RNA products formed are dependent on the oxidants used. Although guanosine was highly susceptible to iron-mediated oxidation, the data in Fig. 1 show that all the ribonucleotides were subjected to oxidative modification with Fe(II)/ascorbate/H2O2 as oxidizing agent. In contrast, guanosine was predominantly oxidized by cyt c/H2O2. However, the level of 8-OH-Guo, which was substantially elevated by the Fenton reaction, was hardly detectable with the cyt c-mediated peroxidation (Fig. 2). Additional products analysis reveals a distinct pattern of oxidized guanosine between the two oxidation reactions. Although it is unknown whether production of 8-OH-Guo is common in heme-mediated peroxidation, we have observed that in some other oxidative reactions, 8-OH-Guo production did not correlate with the oxidative damage (unpublished data). Together, these observations indicate that multiple oxidation markers are necessary for assessing oxidative modification of RNA.
Among the RNA oxidation products is the formation of abasic sites, identified by mass spectrometric methods coupled with liquid chromatography techniques and by its reactivity with the aldehyde-reactive probe. The aldehyde moiety on the abasic sites can readily cross-link with amino group on the protein via Schiff-base formation. In fact, this unique chemical property of abasic sites has been utilized to identify the DNA binding sites on histones and on DNA repair enzymes via in vitro cross-linking between the DNA abasic sites and amino groups of the proteins of interest [35-37]. In addition, Wintermeyer and Zachau [38] took advantage of this chemical property to generate a tRNA probe conjugated with fluorescent dyes by cross-linking to a depuri-nated abasic site under mild reaction conditions. Thus, it is likely that RNA abasic sites generated by cyt c peroxidation could form a cross-linked complex with cyt c. Indeed our results, including those shown in Fig. 5, reveal that RNA and cyt c cross-link in response to peroxidation to form a high-molecular-weight complex that contains both RNA and cyt c. It should be pointed out that cross-linking between ribosomal proteins and rRNA has been reported in yeast induced by H2O2 treatment [21]. In addition, a method has been reported to identify RNA–protein binding sites using UV irradiation to induce the formation of a cross-linked complex between a protein and its bound RNA [39]. These results suggest that substantial amounts of RNA and protein are subjected to cross-linking when they are closely associated under pathological oxidative stress conditions.
What could be the potential physiological function of the RNA–cyt c cross-linked complex? The release of cyt c from the mitochondria to the cytosol is required to initiate the mitochondria-mediated apoptosis [40]. Although the caspase activation pathway initiated by cytoplasmic cyt c has been extensively investigated, it has not been fully delineated how cyt c is released from the mitochondria. Because of its basic properties, cyt c is readily associated with CL, a major acidic lipid in the mitochondrial inner membrane [22,30]. In the presence of an oxidant, heme-containing proteins including cyt c can catalyze peroxidation of a wide variety of organic compounds [41,42]. The peroxidase activity of CL-bound cyt c has been shown to catalyze the oxidation of CL, the only phospholipid in mitochondria that undergoes early oxidation during apoptosis and leads to the release of cyt c [25,26]. Thus, it is hypothesized that peroxidation of the cyt c-bound biomacromolecules may facilitate dissociation of cyt c.
Our results shown in Fig. 7 indicate that treatment of RNA and H2O2 induces cyt c dissociation from the liposomes containing CL in a synergistic manner. Under the experimental conditions, it is reasonable to expect the negatively charged RNA to compete with CL for electrostatic interaction with cyt c. The peroxidation of RNA associated with cyt c forms covalently cross-linked complexes, and as a consequence the population of cyt c-bound CL would be reduced. The results shown in Fig. 7A indicate that cross-linked complexes of cyt c, including those possessing a high-molecular-weight species detected in the supernatant fraction, were elevated as a function of increasing concentration of tRNA and H2O2. However, the majority of cross-linking species, including those generated by cyt c/H2O2-mediated CL oxidation, remained in the liposome fraction (data not shown, also see Fig. 7B). Nevertheless, the results in Figs. 7A and C showed that combination of cyt c, RNA, and H2O2 would not only lead to increasing cyt c-containing cross-linked species, but also facilitate the release of cyt c-containing complexes from the liposomes. Thus, results shown in Fig. 7 suggest that oxidatively induced cyt c release from the CL-containing liposome is mediated by the cyt c-catalyzed per-oxidation of RNA.
If the cyt c-catalyzed RNA peroxidation is involved in oxidative stress-induced cellular apoptosis by facilitating the cyt c release from mitochondria to initiate the caspase activation cascade, RNA has to be accessible in the mitochondrial intermembrane space. Such a possibility is supported by the following findings. (i) Because the mitochondrial membrane transition pore is capable of taking up a molecular size up to 1.5 kDa [43], it is possible that RNA oligomers, resulting from oxidative stress-induced cleavage of microRNA or cytoplasmic tRNAs, could enter into the intermembrane space through the transition pore [44-46]. (ii) Certain cytoplasmic tRNAs are translocated into mammalian mitochondria [47], implying the existence of a natural process to import tRNA into the matrix as well as the intermembrane space. To this end, polynucleotide phosphorylase, a multifunctional ribonuclease, has been located in the intermembrane space [48] and it mediates the translocation of RNAs into mitochondria [49]. In addition, endonuclease G, a nonspecific DNA/RNA nuclease, is located in the intermembrane space under normal conditions [50,51]. These findings together suggest that RNA molecules probably exist in the intermembrane spaces of mitochondria.
To date, RNA oxidation has been shown to exert detrimental physiological effects. For instance, oxidized RNA [52] or RNA containing abasic sites [53] shows inhibitory effects on reverse transcriptase activity, whereas oxidized mRNA [54,55] or mRNA with abasic sites [56] exhibits compromised translation activity as well as translation fidelity. Until now the pathological role of RNA oxidation has been associated only with impaired function of RNA. However, the results of this investigation suggest that oxidative modifications of RNA, including cross-linking, lead not only to impaired RNA normal functions, but also to the gain of a protective signal to facilitate cellular apoptosis in response to oxidative stress.
Acknowledgments
The authors acknowledge the late Dr. Earl R. Stadtman, an eminent scientist and a distinguished mentor, for his initiation and support of this project. This research was supported, in whole, by the Intramural Research Program of the National Institutes of Health; National Heart, Lung, and Blood Institute; and NIH Grants AG11370 and NS056218. Certain commercial equipment or materials are identified in this paper to specify the experimental procedure. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials or equipment identified are necessarily the best available for the purpose.
Abbreviations
- ARP
aldehyde-reactive probe
- CBB
Coomassie brilliant blue
- CL
cardiolipin
- cyt c
cytochrome c
- EDTA
ethylenediaminetetraacetic acid
- GC/MS
gas chromatography–mass spectrometry
- 8-OH-Guo
8-hydroxyguanosine
- 8-OH-Gua
8-hydroxyguanine
- 8-OH-dGuo
8-hydroxydeoxyguanosine
- HPLC
high-performance liquid chromatography
- LC/MS
liquid chromatography–mass spectrometry
- PC
phosphatidylcholine
- ROS
reactive oxygen species
- TFA
trifluoroacetic acid
References
- 1.Stadtman ER, Berlett BS. Reactive oxygen-mediated protein oxidation in aging and disease. Chem Res Toxicol. 1997;10:485–494. doi: 10.1021/tx960133r. [DOI] [PubMed] [Google Scholar]
- 2.Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- 3.Thannickal VJ, Fanburg BL. Reactive oxygen species in cell signaling. Am J Physiol Lung Cell Mol Physiol. 2000;279:L1005–1028. doi: 10.1152/ajplung.2000.279.6.L1005. [DOI] [PubMed] [Google Scholar]
- 4.Evans MD, Dizdaroglu M, Cooke MS. Oxidative DNA damage and disease: induction, repair and significance. Mutat Res. 2004;567:1–61. doi: 10.1016/j.mrrev.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Hofer T, Badouard C, Bajak E, Ravanat JL, Mattsson A, Cotgreave IA. Hydrogen peroxide causes greater oxidation in cellular RNA than in DNA. Biol Chem. 2005;386:333–337. doi: 10.1515/BC.2005.040. [DOI] [PubMed] [Google Scholar]
- 6.Martinet W, de Meyer GR, Herman AG, Kockx MM. Reactive oxygen species induce RNA damage in human atherosclerosis. Eur J Clin Invest. 2004;34:323–327. doi: 10.1111/j.1365-2362.2004.01343.x. [DOI] [PubMed] [Google Scholar]
- 7.Nunomura A, Chiba S, Kosaka K, Takeda A, Castellani RJ, Smith MA, Perry G. Neuronal RNA oxidation is a prominent feature of dementia with Lewy bodies. Neuroreport. 2002;13:2035–2039. doi: 10.1097/00001756-200211150-00009. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, Perry G, Smith MA, Robertson D, Olson SJ, Graham DG, Montine TJ. Parkinson’s disease is associated with oxidative damage to cytoplasmic DNA and RNA in substantia nigra neurons. Am J Pathol. 1999;154:1423–1429. doi: 10.1016/S0002-9440(10)65396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang Y, Kong Q, Shan X, Tian G, Ilieva H, Cleveland DW, Rothstein JD, Borchelt DR, Wong PC, Lin CL. Messenger RNA oxidation occurs early in disease pathogenesis and promotes motor neuron degeneration in ALS. PLoS One. 2008;3:e2849. doi: 10.1371/journal.pone.0002849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nunomura A, Perry G, Hirai K, Aliev G, Takeda A, Chiba S, Smith MA. Neuronal RNA oxidation in Alzheimer’s disease and Down’s syndrome. Ann N Y Acad Sci. 1999;893:362–364. doi: 10.1111/j.1749-6632.1999.tb07855.x. [DOI] [PubMed] [Google Scholar]
- 11.Ding Q, Markesbery WR, Chen Q, Li F, Keller JN. Ribosome dysfunction is an early event in Alzheimer’s disease. J Neurosci. 2005;25:9171–9175. doi: 10.1523/JNEUROSCI.3040-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lovell MA, Markesbery WR. Oxidative damage in mild cognitive impairment and early Alzheimer’s disease. J Neurosci Res. 2007;85:3036–3040. doi: 10.1002/jnr.21346. [DOI] [PubMed] [Google Scholar]
- 13.Markesbery WR, Lovell MA. Damage to lipids, proteins, DNA, and RNA in mild cognitive impairment. Arch Neurol. 2007;64:954–956. doi: 10.1001/archneur.64.7.954. [DOI] [PubMed] [Google Scholar]
- 14.Ide H, Akamatsu K, Kimura Y, Michiue K, Makino K, Asaeda A, Takamori Y, Kubo K. Synthesis and damage specificity of a novel probe for the detection of abasic sites in DNA. Biochemistry. 1993;32:8276–8283. doi: 10.1021/bi00083a031. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka M, Han S, Kupfer PA, Leumann CJ, Sonntag WE. Quantification of oxidized levels of specific RNA species using an aldehyde reactive probe. Anal Biochem. 2011;417:142–148. doi: 10.1016/j.ab.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka M, Han S, Kupfer PA, Leumann CJ, Sonntag WE. An assay for RNA oxidation induced abasic sites using the aldehyde reactive probe. Free Radic Res. 2011;45:237–247. doi: 10.3109/10715762.2010.535529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fornace AJ, Jr, Little JB. DNA–protein cross-linking by chemical carcinogens in mammalian cells. Cancer Res. 1979;39:704–710. [PubMed] [Google Scholar]
- 18.Nackerdien Z, Rao G, Cacciuttolo MA, Gajewski E, Dizdaroglu M. Chemical nature of DNA–protein cross-links produced in mammalian chromatin by hydrogen peroxide in the presence of iron or copper ions. Biochemistry. 1991;30:4873–4879. doi: 10.1021/bi00234a006. [DOI] [PubMed] [Google Scholar]
- 19.Toyokuni S, Mori T, Hiai H, Dizdaroglu M. Treatment of Wistar rats with a renal carcinogen, ferric nitrilotriacetate, causes DNA–protein cross-linking between thymine and tyrosine in their renal chromatin. Int J Cancer. 1995;62:309–313. doi: 10.1002/ijc.2910620313. [DOI] [PubMed] [Google Scholar]
- 20.DeMott MS, Beyret E, Wong D, Bales BC, Hwang JT, Greenberg MM, Demple B. Covalent trapping of human DNA polymerase beta by the oxidative DNA lesion 2-deoxyribonolactone. J Biol Chem. 2002;277:7637–7640. doi: 10.1074/jbc.C100577200. [DOI] [PubMed] [Google Scholar]
- 21.Mirzaei H, Regnier F. Protein–RNA cross-linking in the ribosomes of yeast under oxidative stress. J Proteome Res. 2006;5:3249–3259. doi: 10.1021/pr060337l. [DOI] [PubMed] [Google Scholar]
- 22.Brown LR, Wuthrich K. NMR, ESR studies of the interactions of cyto-chrome c with mixed cardiolipin–phosphatidylcholine vesicles. Biochim Biophys Acta. 1977;468:389–410. doi: 10.1016/0005-2736(77)90290-5. [DOI] [PubMed] [Google Scholar]
- 23.Ding X, Li J, Hu J, Li Q. Electrochemical study of the interaction between cytochrome c and DNA at a modified gold electrode. Anal Biochem. 2005;339:46–53. doi: 10.1016/j.ab.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 24.Mei Y, Yong J, Liu H, Shi Y, Meinkoth J, Dreyfuss G, Yang X. tRNA binds to cytochrome c and inhibits caspase activation. Mol Cell. 2010;37:668–678. doi: 10.1016/j.molcel.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kagan VE, Tyurin VA, Jiang J, Tyurina YY, Ritov VB, Amoscato AA, Osipov AN, Belikova NA, Kapralov AA, Kini V, Vlasova II, Zhao Q, Zou M, Di P, Svistunenko DA, Kurnikov IV, Borisenko GG. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat Chem Biol. 2005;1:223–232. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- 26.Petrosillo G, Ruggiero FM, Paradies G. Role of reactive oxygen species and cardiolipin in the release of cytochrome c from mitochondria. FASEB J. 2003;17:2202–2208. doi: 10.1096/fj.03-0012com. [DOI] [PubMed] [Google Scholar]
- 27.Crain PF. Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry. Methods Enzymol. 1990;193:782–790. doi: 10.1016/0076-6879(90)93450-y. [DOI] [PubMed] [Google Scholar]
- 28.Dizdaroglu M, Rao G, Halliwell B, Gajewski E. Damage to the DNA bases in mammalian chromatin by hydrogen peroxide in the presence of ferric and cupric ions. Arch Biochem Biophys. 1991;285:317–324. doi: 10.1016/0003-9861(91)90366-q. [DOI] [PubMed] [Google Scholar]
- 29.Kupfer PA, Leumann CJ. The chemical stability of abasic RNA compared to abasic DNA. Nucleic Acids Res. 2007;35:58–68. doi: 10.1093/nar/gkl948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rytomaa M, Mustonen P, Kinnunen PK. Reversible, nonionic, and pH-dependent association of cytochrome c with cardiolipin–phosphatidyl-choline liposomes. J Biol Chem. 1992;267:22243–22248. [PubMed] [Google Scholar]
- 31.Babul J, Stellwagen E. Participation of the protein ligands in the folding of cytochrome c. Biochemistry. 1972;11:1195–1200. doi: 10.1021/bi00757a013. [DOI] [PubMed] [Google Scholar]
- 32.Macchioni L, Corazzi T, Davidescu M, Francescangeli E, Roberti R, Corazzi L. Cytochrome c redox state influences the binding and release of cytochrome c in model membranes and in brain mitochondria. Mol Cell Biochem. 2010;341:149–157. doi: 10.1007/s11010-010-0446-1. [DOI] [PubMed] [Google Scholar]
- 33.Hickerson RP, Prat F, Muller JG, Foote CS, Burrows CJ. Sequence and stacking dependence of 8-oxoguanine oxidation: comparison of one-electron vs singlet oxygen mechanisms. J Am Chem Soc. 1999;121:9423–9428. [Google Scholar]
- 34.Pang B, Zhou X, Yu H, Dong M, Taghizadeh K, Wishnok JS, Tannenbaum SR, Dedon PC. Lipid peroxidation dominates the chemistry of DNA adduct formation in a mouse model of inflammation. Carcinogenesis. 2007;28:1807–1813. doi: 10.1093/carcin/bgm037. [DOI] [PubMed] [Google Scholar]
- 35.Deterding LJ, Prasad R, Mullen GP, Wilson SH, Tomer KB. Mapping of the 5′-2-deoxyribose-5-phosphate lyase active site in DNA polymerase beta by mass spectrometry. J Biol Chem. 2000;275:10463–10471. doi: 10.1074/jbc.275.14.10463. [DOI] [PubMed] [Google Scholar]
- 36.Greenberg MM, Weledji YN, Kim J, Bales BC. Repair of oxidized abasic sites by exonuclease III, endonuclease IV, and endonuclease III. Biochemistry. 2004;43:8178–8183. doi: 10.1021/bi0496236. [DOI] [PubMed] [Google Scholar]
- 37.Sun B, Latham KA, Dodson ML, Lloyd RS. Studies on the catalytic mechanism of five DNA glycosylases: probing for enzyme–DNA imino intermediates. J Biol Chem. 1995;270:19501–19508. doi: 10.1074/jbc.270.33.19501. [DOI] [PubMed] [Google Scholar]
- 38.Wintermeyer W, Zachau HG. Replacement of odd bases in tRNA by fluorescent dyes. Methods Enzymol. 1974;29:667–673. doi: 10.1016/0076-6879(74)29058-x. [DOI] [PubMed] [Google Scholar]
- 39.Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein–RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 40.Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 41.Lawrence A, Jones CM, Wardman P, Burkitt MJ. Evidence for the role of a peroxidase compound I-type intermediate in the oxidation of glutathione, NADH, ascorbate, and dichlorofluorescin by cytochrome c/H2O2: implications for oxidative stress during apoptosis. J Biol Chem. 2003;278:29410–29419. doi: 10.1074/jbc.M300054200. [DOI] [PubMed] [Google Scholar]
- 42.Radi R, Thomson L, Rubbo H, Prodanov E. Cytochrome c-catalyzed oxidation of organic molecules by hydrogen peroxide. Arch Biochem Biophys. 1991;288:112–117. doi: 10.1016/0003-9861(91)90171-e. [DOI] [PubMed] [Google Scholar]
- 43.Halestrap AP, McStay GP, Clarke SJ. The permeability transition pore complex: another view. Biochimie. 2002;84:153–166. doi: 10.1016/s0300-9084(02)01375-5. [DOI] [PubMed] [Google Scholar]
- 44.Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583:437–442. doi: 10.1016/j.febslet.2008.12.043. [DOI] [PubMed] [Google Scholar]
- 45.Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rubio MA, Rinehart JJ, Krett B, Duvezin-Caubet S, Reichert AS, Soll D, Alfonzo JD. Mammalian mitochondria have the innate ability to import tRNAs by a mechanism distinct from protein import. Proc Natl Acad Sci USA. 2008;105:9186–9191. doi: 10.1073/pnas.0804283105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen HW, Rainey RN, Balatoni CE, Dawson DW, Troke JJ, Wasiak S, Hong JS, McBride HM, Koehler CM, Teitell MA, French SW. Mammalian polynucleotide phosphorylase is an intermembrane space RNase that maintains mitochondrial homeostasis. Mol Cell Biol. 2006;26:8475–8487. doi: 10.1128/MCB.01002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang G, Chen HW, Oktay Y, Zhang J, Allen EL, Smith GM, Fan KC, Hong JS, French SW, McCaffery JM, Lightowlers RN, Morse HC, 3rd, Koehler CM, Teitell MA. PNPASE regulates RNA import into mitochondria. Cell. 2010;142:456–467. doi: 10.1016/j.cell.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohsato T, Ishihara N, Muta T, Umeda S, Ikeda S, Mihara K, Hamasaki N, Kang D. Mammalian mitochondrial endonuclease G: digestion of R-loops and localization in intermembrane space. Eur J Biochem. 2002;269:5765–5770. doi: 10.1046/j.1432-1033.2002.03238.x. [DOI] [PubMed] [Google Scholar]
- 51.Schafer P, Scholz SR, Gimadutdinow O, Cymerman IA, Bujnicki JM, Ruiz-Carrillo A, Pingoud A, Meiss G. Structural and functional characterization of mitochondrial EndoG, a sugar non-specific nuclease which plays an important role during apoptosis. J Mol Biol. 2004;338:217–228. doi: 10.1016/j.jmb.2004.02.069. [DOI] [PubMed] [Google Scholar]
- 52.Rhee Y, Valentine MR, Termini J. Oxidative base damage in RNA detected by reverse transcriptase. Nucleic Acids Res. 1995;23:3275–3282. doi: 10.1093/nar/23.16.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kupfer PA, Crey-Desbiolles C, Leumann CJ. Trans-lesion synthesis and RNaseH activity by reverse transcriptases on a true abasic RNA template. Nucleic Acids Res. 2007;35:6846–6853. doi: 10.1093/nar/gkm767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shan X, Chang Y, Lin CL. Messenger RNA oxidation is an early event preceding cell death and causes reduced protein expression. FASEB J. 2007;21:2753–2764. doi: 10.1096/fj.07-8200com. [DOI] [PubMed] [Google Scholar]
- 55.Tanaka M, Chock PB, Stadtman ER. Oxidized messenger RNA induces translation errors. Proc Natl Acad Sci USA. 2007;104:66–71. doi: 10.1073/pnas.0609737104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hudak KA, Bauman JD, Tumer NE. Pokeweed antiviral protein binds to the cap structure of eukaryotic mRNA and depurinates the mRNA downstream of the cap. RNA. 2002;8:1148–1159. doi: 10.1017/s1355838202026638. [DOI] [PMC free article] [PubMed] [Google Scholar]


