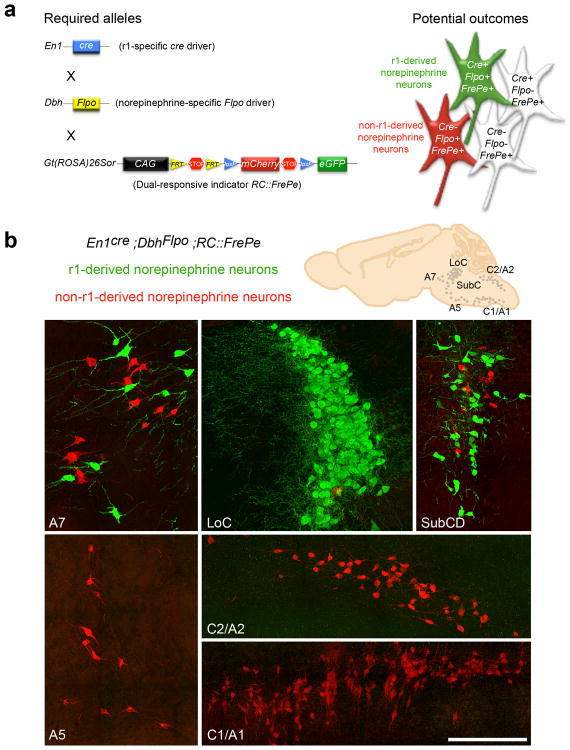
Fig. 1. Intersectional genetic fate mapping strategy distinguishes r1(En1cre)-derived from non-r1-derived norepinephrine neurons.
(a) Visualization of r1-derived norepinephrine neurons in isolation requires a r1-specific cre driver line (En1cre), a norepinephrine-specific Flpo driver line (DbhFlpo, see Supplementary Fig. 1), and a dual recombinase-responsive indicator line (RC∷FrePe). In mice that inherit all three alleles, norepinephrine neurons from r1 will express both Flpo and cre resulting in eGFP expression (green neuron in schematic), and all non-r1-derived norepinephrine neurons will express only Flpo resulting in mCherry expression (red neuron in schematic). Non-norepinephrine neurons will not express Flpo and are not marked by a fluorophore regardless of Cre expression (white cells in schematic). (b) Sections from En1Cre;DbhFlpo;RC∷FrePe adult mouse brainstem immunostained for eGFP and mCherry reveal the contribution of r1(En1cre)-derived norepinephrine neurons (green) to A7, LoC, and SubCD nuclei, and non-r1-derived norepinephrine neurons (red) to A7, LoC, SubCD, A5, C2/A2 and C1/A1 nuclei. Scale bar indicates 200 μm (A7, LoC, SubCD, and A5 coronal images) and 218 μm (C2/A2 and C1/A1 sagittal images).