Abstract
Francisella tularensis, the agent of the zoonotic disease tularemia, is a highly infectious bacterium for a large number of animal species and can be transmitted to humans by various means. The bacterium is able to infect a variety of cell types but replicates in mammalian hosts mainly in the cytosol of infected macrophages. In order to resist the stressful and nutrient-restricted intracellular environments, it encounters during its systemic dissemination, Francisella has developed dedicated stress resistance mechanisms and adapted its metabolic and nutritional needs. Recent data form our laboratory and from several other groups have shown that Francisella simultaneously relies on multiple host amino acid sources during its intracellular life cycle. This review will summarize how intracellular Francisella use different amino acid sources, and their role in phagosomal escape and/or cytosolic multiplication and systemic dissemination. We will first summarize the data that we have obtained on two amino acid transporters involved in Francisella phagosomal escape and cytosolic multiplication i.e., the glutamate transporter GadC and the asparagine transporter AnsP, respectively. The specific contribution of glutamate and asparagine to the physiology of the bacterium will be evoked. Then, we will discuss how Francisella has adapted to obtain and utilize host amino acid resources, and notably the contribution of host transporters and autophagy process in the establishment of a nutrient-replete intracellular niche.
Keywords: nutrition, intracellular pathogen, amino acid uptake, phagosomal escape, Francisella
Introduction
Francisella tularensis is a small Gram-negative bacillus, aerobic, non-spore-forming, and non-motile. This facultative intracellular pathogen is the causative agent of the zoonotic disease tularemia in a large number of animal species. This highly infectious bacterial pathogen can be transmitted to humans in numerous ways (Sjostedt, 2011), including direct contact with sick animals, inhalation, ingestion of contaminated water or food, or by bites from ticks, mosquitoes or flies. Four different subspecies (subsp.) of F. tularensis that differ in virulence and geographic distribution exist, designated subsps. tularensis, holarctica, mediasiatica, and novicida, respectively. The most virulent subspecies tularensis is considered a potential Class A agent in bioterrorism by the Centers for Disease Control (CDC) in the United States (Oyston et al., 2004; Keim et al., 2007). F. tularensis subsp. novicida (F. novicida) is rarely pathogenic to non-immunocompromized humans but is fully virulent for mice and is therefore widely used as a model to study Francisella intracellular parasitism.
F. novicida has the capacity to evade host defenses and to replicate to high numbers within the cytosol of eukaryotic cells (Jones et al., 2012). The bacterium is able to replicate inside a variety of cells, and in particular in macrophages. After a transient passage through a phagosomal compartment, bacteria are released within 30–60 min in the host cell cytosol where they undergo several rounds of active replication (Celli and Zahrt, 2013). At least 20% of the genome participates to some extent to Francisella virulence (Meibom and Charbit, 2010b), including an important proportion of genes related to metabolic and nutritional functions. However, understanding the relationship between nutrition and the in vivo life cycle of Francisella is still poorly understood.
Francisella is predicted to possess numerous nutrient uptake systems to capture its necessary host-derived nutrients, some of which are probably available in limiting concentrations. We will review here our recent findings regarding two Francisella amino acid acquisition systems and their importance in the physiology and intracellular life cycle of Francisella. We will also discuss the major host responses, identified to date, triggered upon Francisella infection that contribute to fuel the cytosolic compartment.
Francisella amino acid transporters play a critical role in intracellular multiplication
We have previously shown that Francisella used the cysteine-containing tripeptide glutathione (GSH) as a source of cysteine, to replicate in infected macrophages (Alkhuder et al., 2009), thus suggesting that this bacterium has evolved by exploiting the natural abundance of GSH in the host cytosol to compensate its natural auxotrophy for cysteine. More recently, we decided to evaluate the role of amino acid transport systems in the capacity of Francisella to thrive intracellularly.
As mentioned in an earlier review (Meibom and Charbit, 2010a), most of the transport systems encoded by Francisella genomes are secondary carriers, corresponding to up to 75% of the predicted transport proteins in F. tularensis. These active transport systems use the electrochemical gradient to translocate elements across the bacterial cytoplasmic membrane, either by symport (two components moving in the same direction) or by antiport (two molecules moving in opposite direction). Secondary transporters of Francisella encompass several major families such as: (i) the major facilitator superfamily (MFS), involved in various functions, including drug efflux, sugar and amino acid uptake (Meibom and Charbit, 2010a), and comprising 31 proteins; (ii) the amino acid/polyamine/organocation (APC) subfamily, comprising 11 proteins; (iii) the subfamily of permeases for hydroxy aromatic amino acids (HAAAP), comprising 7 proteins; and (iv) the oligopeptide dependent proton (POT) subfamily of transporters, comprizing 8 proteins. Remarkably, 52 proteins involved in secondary active transport were found in earlier in vitro or in vivo genetic screens as being involved in Francisella pathogenicity (Qin and Mann, 2006; Tempel et al., 2006; Maier et al., 2007; Su et al., 2007; Weiss et al., 2007; Kraemer et al., 2009; Asare and Abu Kwaik, 2010; Asare et al., 2010; Moule et al., 2010; Peng and Monack, 2010), among which 61% of the MFS transporters.
We will discuss below the data that we have obtained very recently on GadC and AnsP, two secondary transporters being specifically involved in phagosomal escape and cytosolic multiplication, respectively.
The APC family members
F. tularensis genomes encode 11 predicted members of the amino acid-polyamine-organocation (APC) superfamily transporters, specifically involved in amino acids exchange w/o ions. Remarkably, 8 of the 11 APC members have been identified at least once in earlier genetic studies (in vitro or in vivo). The functional role of one of them, the glutamate transporter GadC, was elucidated in our laboratory (Ramond et al., 2014) and its essential contribution to Francisella phagosomal escape was demonstrated.
Glutamate uptake is critical in the phagosome
The GadC-encoding gene (FTN_0571 in F. novicida) had been identified in four studies as an essential actor in F. tularensis subsp. holarctica (Maier et al., 2007) and F. novicida (Weiss et al., 2007; Kraemer et al., 2009; Peng and Monack, 2010) virulence. Inactivation of the gene FTN_0571 in F. novicida confirmed its implication in phagocytic cells multiplication such as J774.1, THP-1 and bone marrow-derived macrophages (BMMs) but also in virulence (in BALB/c mice). We showed that this transporter was essential for proper bacterial phagosomal escape. Furthermore, our analyses revealed that GadC contributed to the resistance to reactive oxygen species (ROS) generated in the phagosome by importing glutamate. Remarkably, the intracellular multiplication defect, as well as attenuated virulence in mice of the ΔgadC mutant, was fully or partially reversed in phox-KO BMMs and phox-KO mice, respectively. Altogether, our data were consistent with the notion that imported glutamate was used by F. tularensis to fuel the tricarboxylic acid cycle (TCA) in the phagosomal compartment, revealing thus a new role of glutamate utilization in oxidative stress defense. Of note, GadC was previously shown to play a specific role in acid resistance challenge, especially in E. coli (Castanie-Cornet et al., 1999) and L. monocytogenes (Cotter et al., 2001), two enteropathogenic bacteria that must resist a severe acid stress during their natural infectious route. The role of GadC in oxidative stress resistance in F. tularensis, which is not an enteropathogen, may thus, illustrate an adaptation of bacterium to the environments it encounters during its in vivo life cycle.
The MFS transporters
The importance of the MFS family in the virulence of intracellular bacteria has been demonstrated for the first time in Legionella pneumophila. Indeed, a threonine transporter (called PhtA for Phagosomal Transporter A) has been identified as unable to differentiate into replicative form in macrophages and as expressing an early stage of the factors leading to the transmissive form in culture medium (Sauer et al., 2005). The L. pneumophila genome encodes 10 additional PhtA paralogues (Sauer et al., 2005), some of which are also required during intracellular replication (Fonseca and Swanson, 2014). PhtJ is required for acquisition of valine and PhtC and PhtD were very recently shown to contribute in protecting L. pneumophila from dTMP starvation (Fonseca et al., 2014).
Strikingly, Pht transporters constitute a sub-family of MFS transporters found exclusively in intracellular pathogens and specifically in Alpha and Gamma-proteobacteria (including L. pneumophila, Coxiella burnetii, Rickettsiella grylli, Francisella tularensis, Wolbachia, Anaplasma, Ehrlichia, Protochlamidia amoebophila, Prochlorococcus marinus, and Zymomonas mobilis (Chen et al., 2008).
This led us to address in priority the role of the Pht subfamily members in Francisella pathogenesis. The six Pht transporters identified in Francisella are highly conserved in the different sub-species (>95% amino acid identity). Each of these transporters has been found to contribute to F. tularensis virulence (Qin and Mann, 2006; Weiss et al., 2007; Kraemer et al., 2009; Asare and Abu Kwaik, 2010; Asare and Kwaik, 2010; Moule et al., 2010; Peng and Monack, 2010; Llewellyn et al., 2011).
The functional role of two Pht transporters of F. tularensis was very recently elucidated in our laboratory (Gesbert et al., 2014, 2015). We will focus below on one of them, the asparagine transporter AnsP that contributes exclusively to the cytosolic multiplication of the pathogen.
Asparagine uptake is critical for cytosolic multiplication
Inactivation of the ansP gene in F. tularensis subsp tularensis SCHU S4, F. tularensis subsp holarctica LVS or F. tularensis subsp novicida U112, causes a decrease in intracellular multiplication in different cellular models such as HepG2 human hepatocytes, primary murine macrophages and murine macrophages J774 (Qin and Mann, 2006; Marohn et al., 2012; Gesbert et al., 2014). In vivo, the inactivation of the ansP gene causes a significant decrease in virulence of F. tularensis subsp novicida U112 in Drosophila (Asare and Abu Kwaik, 2010); and F. tularensis subsp holarctica LVS in BALB/c mice (Marohn et al., 2012).
We showed that the impaired intracellular growth and the F. novicida ΔansP mutant could be abrogated upon supplementation with an excess of asparagine: (i) in a free form or as a dipeptide, in cell culture models; (ii) by intraperitoneal injection of asparagine, in infected mice (Gesbert et al., 2014). Thus, whereas the bacterium is prototrophic for asparagine during growth in synthetic medium, it becomes phenotypically auxotrophic for asparagine in the cytosol of infected cells. A similar phenomenon has been also described in M. tuberculosis. This bacterium, which is prototrophic in culture for the 20 amino acids, possesses two transporters, one for aspartate and one for asparagine, which are important for intracellular mycobacterial survival and multiplication (Gouzy et al., 2013, 2014). Thus, the existence of a predicted active biosynthetic pathway does not preclude the requirement of an efficient transport system to support intracellular multiplication. These results support the generally accepted assumption that nutrient uptake is favored to synthesis when the nutrient is available in the external medium. We also demonstrated that the AnsP-mediated capture of asparagine by Francisella had no impact on phagosomal escape but became critical as soon as the bacterium reached the host cytoplasm (Gesbert et al., 2014). The data obtained further suggested that cytosolic bacteria use asparagine mainly as a building block for protein synthesis. However, additional metabolic or regulatory functions of asparagine cannot be excluded.
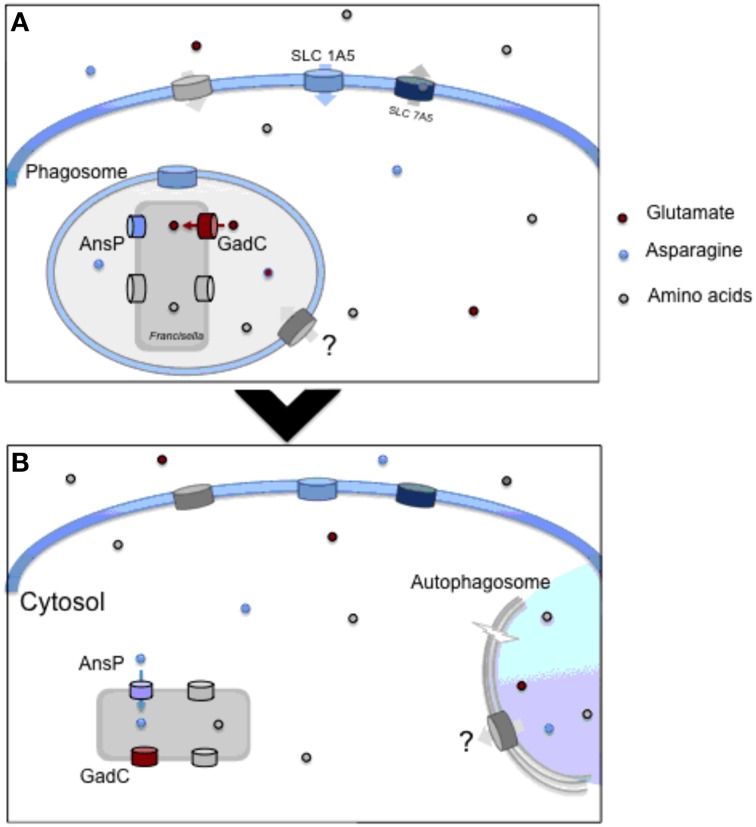
It is likely that other members of the MFS and APC families (and possibly of other families) yet to be discovered, may participate to the intracellular life cycle of Francisella (Figure 1).
Figure 1.
Amino acid utilzation by intracellular Francisella. (A) Upon entry into macrophages, Francisella transiently resides in a phagosomal compartment. In this compartment, the glutamate permease GadC helps the bacterium to resist the oxidative stress generated by the NADPH oxidase. This transporter is critically required for proper bacterial phagosomal escape. Concomitantly, the synthesis of the host neutral amino acid transporter SLC1A5 is rapidly up-regulated, favoring the entry of amino acids in the infected host cell. (B) Once in the host cytosol, bacteria start their active replication and therefore require the supply of numerous additional host-derived nutrients. The AnsP permease contributes to bacterial multiplication by providing asparagine for protein synthesis. The ATG-5-independent autophagy contributes to the delivery of amino acids, enriching the cytosolic bacterial diet. At both stages, other amino acid transporters contribute to the proper feeding of the bacterium.
Host-derived amino acid sources
The host cytosol, previously considered as a safe nutrient-replete (Ray et al., 2009), is now established as a life-threatening nutrient-deprived environment for the invading bacteria (Abu Kwaik and Bumann, 2013). A permanent war takes place between the host and the pathogen. Indeed, intracellular pathogens try to deprive nutrients from their host, while the host cell tries to deprive the invading bacteria of nutrients. This phenomenon is generally designated “nutritional immunity” (Barel and Charbit, 2013).
Host amino acid transporters
Involvement of a eukaryotic glutamine transporter SLC1A5 has been recently described during F. tularensis LVS infection (Barel et al., 2012). F. tularensis LVS induces up-regulation of this transporter both at the mRNA and the protein level. Furthermore, the bacteria infection induces the deglycosylation of this aminoacid transporter. This deglycosylation process increases with the time of infection and is correlated with the increase in SLC1A5 expression. The IglC mutant, that does not egress from the phagosome and does not muliply in the cytosol, does not induce this deglycosylation. Therefore, this deglycosylation is induced only by bacteria, which are able to evade the phagosome and multiply into the cytosol.
In the same time, SLC7A5 mRNA and protein expression is down-regulated. SLC7A5 is a partner of SLC1A5 as they work along to equilibrate the cytoplasmic amino acid pool (Fuchs and Bode, 2005) and especially of glutamine (Gln). The differential effect of F. tularensis LVS infection on SLC1A5/SLC7A5 expression could therefore induce an increase in intracellular concentration of glutamine. This nutrient plays an important role in regulating gene expression, protein turnover, anti-oxidative function, nutrient metabolism, immunity, and acid-base balance. Use of glutamine by human cells, for controlling Francisella infection remains to be studied. We have found (Barel and Charbit, 2013) that addition of glutamine increased the ability of F. tularensis LVS to multiply in the cytosol of infected THP-1 cells. Although classified as “non-essential,” glutamine appears essential for the viability and growth of cells maintained in tissue culture by serving notably as a metabolic precursor in several biosynthetic pathways, or directly for protein synthesis (Neu et al., 1996). Glutamine deprivation mediated through Helicobacter bilis γ-Glutamyl-transpeptidase has been found to be responsible for induction of inflammatory disorders in epithelial cells (Javed et al., 2013).
Of note, SLC1A5 gene is activated by insulin, through SGK1, SGK3, and PKB kinase activation, which stimulates amino acid absorption (Palmada et al., 2005). In good relationship, we indeed found that addition of insulin also increased F. tularensis LVS intracellular multiplication (Barel et al., 2012). Glutamine may be converted into glutamate that in turn, can be used either by the bacteria or by the human cells to provide metabolic advantages and prevent Francisella infection. As discussed above, the glutamate transporter of Francisella (GadC) has been shown to be critical for oxidative stress defense in the phagosome (Ramond et al., 2014).
The deglycosylation of SLC1A5 seems to be a general mechanism induced by F. tularensis infection, as deglycosylation of another highly glycosylated protein (CD147) was also observed. Protein deglycosylation triggered by intracellular Francisella may therefore serve also as a nutrient source. The glycan produced could serve in networks regulating cellular involvement directed toward survival of intracellular bacteria.
Autophagy
Utilization of the autophagy pathway has recently been shown to constitute another mechanism of bacterial adaptation, contributing to the survival and nutrition of intracellular Francisella (Steele et al., 2013). Interestingly, Kawula and co-worker have found that intracellular F. tularensis subsp tularensis SCHU S4 relied on ATG5-independent autophagy for multiplication. Supplying excess pyruvate or amino acids suppressed the bacterial growth defect in autophagy-deficient cells, suggesting a direct role of this process in supplying amino acid. However, F. tularensis subsp holarctica LVS intracellular growth appeared to be significantly impaired in ATG5−/− macrophages. It is thus likely that this subspecies may use both ATG5-dependent and ATG5-independent autophagy processes to support efficient intracellular growth. Altogether, these data indicate that wild-type intracellular F. tularensis generally avoid engulfment by classical autophagosomes but may use autophagy to fuel cytosolic bacterial replication with amino acid. It is possible that other nutrients are provided to intracellular bacteria by this process (such as carbohydrates or lipids).
These two hosts responses triggered upon infection reflect the capacity of intracellular Francisella to utilize the eukaryotic machinery for producing amino acids and glycans residues, which may serve for its own survival.
Concluding remarks
We have shown that two Francisella transporters, involved in the uptake of asparagine and glutamate, played a critical role in the bacterial intracellular life cycle. Remarkably, these two amino acids are two non-essential amino acids, thus implying that prototrophy in broth does not necessarily predict independence toward host amino acid source(s) during infection. Comparison of predicted nutrient utilization and biosynthetic pathways of a series of mammalian pathogens support the notion that most pathogens share the capability to simultaneously utilize multiple nitrogen and carbon sources (Abu Kwaik and Bumann, 2013; Steeb et al., 2013). Our ongoing studies indicate that Francisella also relies on several other host-derived amino acid sources to multiply inside infected macrophages (Gesbert et al., 2015, unpublished).
Other host nutrient sources are likely to be involved in Francisella intracellular adaptation. For example, Weiss and co-workers have shown that biotin biosynthesis was required in the Francisella phagosome to promote rapid bacterial escape (Napier et al., 2012), providing the first example of a metabolic requirement of Francisella in this compartment. The importance of biotin biogenesis in phagosomal escape may suggest that biotin is limiting in this compartment. If so, sequestration of biotin could constitute a form of nutritional immunity by the host innate immune system. These hypothesis remains to be experimentally confirmed. Iron is also limiting in the phagosome and numerous host factors play a critical role in the control of iron sequestration upon microbial infection. F. tularensis subsp holarctica LVS infection has been shown to trigger a significant up-regulation of transferrin receptor expression, ultimately leading to increased intracellular iron availability. However, the host combats this iron influx by expelling iron out of the phagosomes as well as by increasing the expression ferroportin, which shuttles iron from the cytosol out of the cell. In turn, to reduce ferroortin activity, F. tularensis subsp holarctica LVS infection induces the production of hepcidin that binds to ferroportin on the host cell surface, leading to its subsequent degradation (Pan et al., 2010; Jones et al., 2012). As a result, Francisella is able to maintain a sufficient intracellular iron pool.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
These studies were supported by INSERM, CNRS and Université Paris Descartes Paris Cité Sorbonne. Gael Gesbert was funded by a fellowship from the “Délégation Générale à l'Armement” (DGA) and Elodie Ramond by a fellowship from the “Région Ile de France.”
References
- Abu Kwaik Y., Bumann D. (2013). Microbial quest for food in vivo: ‘nutritional virulence’ as an emerging paradigm. Cell. Microbiol. 15, 882–890. 10.1111/cmi.12138 [DOI] [PubMed] [Google Scholar]
- Alkhuder K., Meibom K. L., Dubail I., Dupuis M., Charbit A. (2009). Glutathione provides a source of cysteine essential for intracellular multiplication of Francisella tularensis. PLoS Pathog. 5:e1000284. 10.1371/journal.ppat.1000284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asare R., Abu Kwaik Y. (2010). Molecular complexity orchestrates modulation of phagosome biogenesis and escape to the cytosol of macrophages by Francisella tularensis. Environ. Microbiol. 12, 2559–2586. 10.1111/j.1462-2920.2010.02229.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asare R., Akimana C., Jones S., Abu Kwaik Y. (2010). Molecular bases of proliferation of Francisella tularensis in arthropod vectors. Environ. Microbiol. 12, 2587–2612. 10.1111/j.1462-2920.2010.02230.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asare R., Kwaik Y. A. (2010). Exploitation of host cell biology and evasion of immunity by Francisella tularensis. Front. Microbiol. 1:145. 10.3389/fmicb.2010.00145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barel M., Charbit A. (2013). Francisella tularensis intracellular survival: to eat or to die. Microbes Infect. 15, 989–997 10.1016/j.micinf.2013.09.009 [DOI] [PubMed] [Google Scholar]
- Barel M., Meibom K., Dubail I., Botella J., Charbit A. (2012). Francisella tularensis regulates the expression of the amino acid transporter SLC1A5 in infected THP-1 human monocytes. Cell. Microbiol. 14, 1769–1783. 10.1111/j.1462-5822.2012.01837.x [DOI] [PubMed] [Google Scholar]
- Castanie-Cornet M. P., Penfound T. A., Smith D., Elliott J. F., Foster J. W. (1999). Control of acid resistance in Escherichia coli. J. Bacteriol. 181, 3525–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli J., Zahrt T. C. (2013). Mechanisms of Francisella tularensis intracellular pathogenesis. Cold Spring Harb. Perspect. Med. 3:a010314. 10.1101/cshperspect.a010314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D. E., Podell S., Sauer J. D., Swanson M. S., Saier M. H., Jr. (2008). The phagosomal nutrient transporter (Pht) family. Microbiology 154, 42–53. 10.1099/mic.0.2007/010611-0 [DOI] [PubMed] [Google Scholar]
- Cotter P. D., O'Reilly K., Hill C. (2001). Role of the glutamate decarboxylase acid resistance system in the survival of Listeria monocytogenes LO28 in low pH foods. J. Food Prot. 64, 1362–1368. [DOI] [PubMed] [Google Scholar]
- Fonseca M. V., Sauer J. D., Crepin S., Byrne B., Swanson M. S. (2014). The phtC-phtD Locus Equips Legionella pneumophila for Thymidine Salvage and Replication in Macrophages. Infect. Immun. 82, 720–730. 10.1128/IAI.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca M. V., Swanson M. S. (2014). Nutrient salvaging and metabolism by the intracellular pathogen Legionella pneumophila. Front. Cell. Infect. Microbiol. 4:12. 10.3389/fcimb.2014.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs B. C., Bode B. P. (2005). Amino acid transporters ASCT2 and LAT1 in cancer: partners in crime? Semin. Cancer Biol. 15, 254–266. 10.1016/j.semcancer.2005.04.005 [DOI] [PubMed] [Google Scholar]
- Gesbert G., Ramond E., Rigard M., Frapy E., Dupuis M., Dubail I., et al. (2014). Asparagine assimilation is critical for intracellular replication and dissemination of Francisella. Cell. Microbiol. 16, 434–449. 10.1111/cmi.12227 [DOI] [PubMed] [Google Scholar]
- Gesbert G., Ramond E., Tros F., Dairou J., Frapy E., Barel M., et al. (2015). Importance of branched-chain amino acid utilization in Francisella intracellular adaptation. Infect Immun. 83, 173–183. 10.1128/IAI.02579-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouzy A., Larrouy-Maumus G., Bottai D., Levillain F., Dumas A., Wallach J., et al. (2014). Mycobacterium tuberculosis exploits asparagine to assimilate nitrogen and resist acid stress during infection. PLoS Pathog. 10:e1003928. 10.1371/journal.ppat.1003928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouzy A., Larrouy-Maumus G., Wu T. D., Peixoto A., Levillain F., Lugo-Villarino G., et al. (2013). Mycobacterium tuberculosis nitrogen assimilation and host colonization require aspartate. Nat. Chem. Biol. 9, 674–676. 10.1038/nchembio.1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed S., Mejias-Luque R., Kalali B., Bolz C., Gerhard M. (2013). Helicobacter bilis gamma-glutamyltranspeptidase enhances inflammatory stress response via oxidative stress in colon epithelial cells. PLoS ONE 8:e73160. 10.1371/journal.pone.0073160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. L., Napier B. A., Sampson T. R., Llewellyn A. C., Schroeder M. R., Weiss D. S. (2012). Subversion of host recognition and defense systems by Francisella spp. Microbiol. Mol. Biol. Rev. 76, 383–404. 10.1128/MMBR.05027-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim P., Johansson A., Wagner D. M. (2007). Molecular epidemiolgy, evolution, and ecology of Francisella. Ann. N.Y. Acad. Sci. 1105, 30–66. 10.1196/annals.1409.011 [DOI] [PubMed] [Google Scholar]
- Kraemer P. S., Mitchell A., Pelletier M. R., Gallagher L. A., Wasnick M., Rohmer L., et al. (2009). Genome-wide screen in Francisella novicida for genes required for pulmonary and systemic infection in mice. Infect. Immun. 77, 232–244. 10.1128/IAI.00978-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn A. C., Jones C. L., Napier B. A., Bina J. E., Weiss D. S. (2011). Macrophage replication screen identifies a novel Francisella hydroperoxide resistance protein involved in virulence. PLoS ONE 6:e24201. 10.1371/journal.pone.0024201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier T. M., Casey M. S., Becker R. H., Dorsey C. W., Glass E. M., Maltsev N., et al. (2007). Identification of Francisella tularensis Himar1-based transposon mutants defective for replication in macrophages. Infect. Immun. 75, 5376–5389. 10.1128/IAI.00238-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marohn M. E., Santiago A. E., Shirey K. A., Lipsky M., Vogel S. N., Barry E. M. (2012). Members of the Francisella tularensis phagosomal transporter subfamily of major facilitator superfamily transporters are critical for pathogenesis. Infect. Immun. 80, 2390–2401. 10.1128/IAI.00144-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meibom K. L., Charbit A. (2010a). Francisella tularensis metabolism and its relation to virulence. Front. Microbiol. 1:140. 10.3389/fmicb.2010.00140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meibom K. L., Charbit A. (2010b). The unraveling panoply of Francisella tularensis virulence attributes. Curr. Opin. Microbiol. 13, 11–17. 10.1016/j.mib.2009.11.007 [DOI] [PubMed] [Google Scholar]
- Moule M. G., Monack D. M., Schneider D. S. (2010). Reciprocal analysis of Francisella novicida infections of a Drosophila melanogaster model reveal host-pathogen conflicts mediated by reactive oxygen and imd-regulated innate immune response. PLoS Pathog. 6:e1001065. 10.1371/journal.ppat.1001065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napier B. A., Meyer L., Bina J. E., Miller M. A., Sjostedt A., Weiss D. S. (2012). Link between intraphagosomal biotin and rapid phagosomal escape in Francisella. Proc. Natl. Acad. Sci. U.S.A. 109, 18084–18089. 10.1073/pnas.1206411109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu J., Shenoy V., Chakrabarti R. (1996). Glutamine nutrition and metabolism: where do we go from here? FASEB J. 10, 829–837. [DOI] [PubMed] [Google Scholar]
- Oyston P. C., Sjostedt A., Titball R. W. (2004). Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat. Rev. Microbiol. 2, 967–978. 10.1038/nrmicro1045 [DOI] [PubMed] [Google Scholar]
- Palmada M., Speil A., Jeyaraj S., Bohmer C., Lang F. (2005). The serine/threonine kinases SGK1, 3 and PKB stimulate the amino acid transporter ASCT2. Biochem. Biophys. Res. Commun. 331, 272–277. 10.1016/j.bbrc.2005.03.159 [DOI] [PubMed] [Google Scholar]
- Pan X., Tamilselvam B., Hansen E. J., Daefler S. (2010). Modulation of iron homeostasis in macrophages by bacterial intracellular pathogens. BMC Microbiol. 10:64. 10.1186/1471-2180-10-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng K., Monack D. M. (2010). Indoleamine 2,3-dioxygenase 1 is a lung-specific innate immune defense mechanism that inhibits growth of Francisella tularensis tryptophan auxotrophs. Infect. Immun. 78, 2723–2733. 10.1128/IAI.00008-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin A., Mann B. J. (2006). Identification of transposon insertion mutants of Francisella tularensis tularensis strain Schu S4 deficient in intracellular replication in the hepatic cell line HepG2. BMC Microbiol. 6:69. 10.1186/1471-2180-6-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramond E., Gesbert G., Rigard M., Dairou J., Dupuis M., Dubail I., et al. (2014). Glutamate utilization couples oxidative stress defense and the tricarboxylic acid cycle in Francisella phagosomal escape. PLoS Pathog. 10:e1003893. 10.1371/journal.ppat.1003893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K., Marteyn B., Sansonetti P. J., Tang C. M. (2009). Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat. Rev. Microbiol. 7, 333–340. 10.1038/nrmicro2112 [DOI] [PubMed] [Google Scholar]
- Sauer J. D., Bachman M. A., Swanson M. S. (2005). The phagosomal transporter A couples threonine acquisition to differentiation and replication of Legionella pneumophila in macrophages. Proc. Natl. Acad. Sci. U.S.A. 102, 9924–9929. 10.1073/pnas.0502767102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjostedt A. (2011). Special Topic on Francisella tularensis and Tularemia. Front. Microbiol. 2:86. 10.3389/fmicb.2011.00086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeb B., Claudi B., Burton N. A., Tienz P., Schmidt A., Farhan H., et al. (2013). Parallel exploitation of diverse host nutrients enhances Salmonella virulence. PLoS Pathog. 9:e1003301. 10.1371/journal.ppat.1003301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele S., Brunton J., Ziehr B., Taft-Benz S., Moorman N., Kawula T. (2013). Francisella tularensis harvests nutrients derived via ATG5-independent autophagy to support intracellular growth. PLoS Pathog. 9:e1003562. 10.1371/journal.ppat.1003562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J., Yang J., Zhao D., Kawula T. H., Banas J. A., Zhang J.-R. (2007). Genome-wide identification of Francisella tularensis virulence determinants. Infect. Immun. 75, 3089–3101. 10.1128/IAI.01865-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempel R., Lai X. H., Crosa L., Kozlowicz B., Heffron F. (2006). Attenuated Francisella novicida transposon mutants protect mice against wild-type challenge. Infect. Immun. 74, 5095–5105. 10.1128/IAI.00598-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D. S., Brotcke A., Henry T., Margolis J. J., Chan K., Monack D. M. (2007). In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl. Acad. Sci. U.S.A. 104, 6037–6042. 10.1073/pnas.0609675104 [DOI] [PMC free article] [PubMed] [Google Scholar]