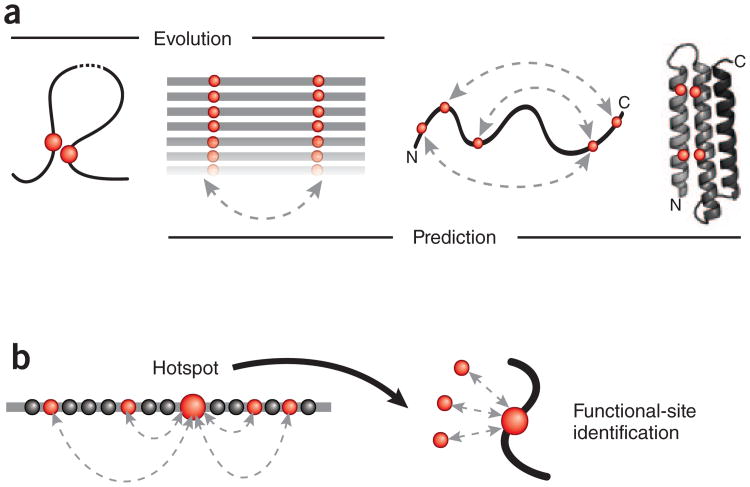
Figure 1.
Reading the sequence record for evolutionary constraints. (a) Evolutionary pressure (left) to maintain favorable interactions between physically interacting amino acid residues (red circles) in the three-dimensional fold of a protein (curved line) leaves a visible record of residue covariation (double-headed, dashed arrow) in related protein sequences (aligned horizontal lines). The inverse problem of inferring (right) directly causative residue couplings (evolutionary couplings) from the covariation record is challenging because of transitive correlations and other confounding effects, but once evolutionary couplings are determined (double-headed dashed arrows on curved protein chain), they can be used to predict the unknown three-dimensional structure of a protein (ribbon, right) from a set of sequences alone. (b) Residues subject to a high number of evolutionary pair constraints (double-headed, dashed arrows; left) represent likely functional hotspots (large red dot). Such highly constrained residues include residues in functional sites (for example, interaction with external ligands, red dots on right) that may not be detectable by analysis of single-residue conservation.