Figure 3.
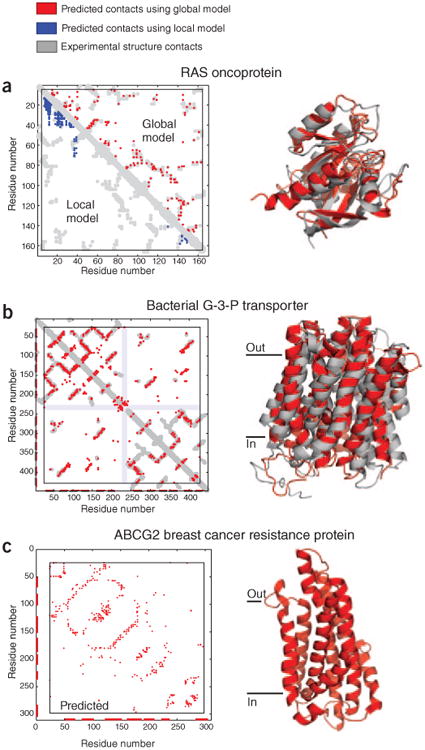
High-ranking evolutionary constraints correspond well to experimental structure contacts in blinded tests, encouraging prediction of unknown structures. (a) Blinded prediction test for a globular protein. Dots in plots on left represent contacts between residues in a protein. Residue pairs with high coevolution scores from local models based on mutual information are mostly not close in three dimensions (blue dots), whereas high-ranking evolutionary constraints (red dots) correspond well to experimental structure contacts (gray). The same number of predictions are shown in each triangle (same number of blue and red dots). The high accuracy of prediction of evolutionary constraints allows the prediction of the all-atom three-dimensional structures of globular proteins, shown as a ribbon diagram of the human oncoprotein RAS (red, evolutionary coupling–based prediction; gray, crystal structure; Uniprot identifier RASH_HUMAN; PDB identifier 5p21)15. (b) Blinded prediction test as in a for a transmembrane protein (Uniprot identifier GLPT_ECOLI; PDB identifier, 1pw4 (ref. 22). (c) Example of prediction of a medically important protein of unknown three-dimensional structure, ATP-binding cassette sub-family G member 2 (alias, breast cancer resistance protein, Uniprot identifier ABCG2_HUMAN)22.
