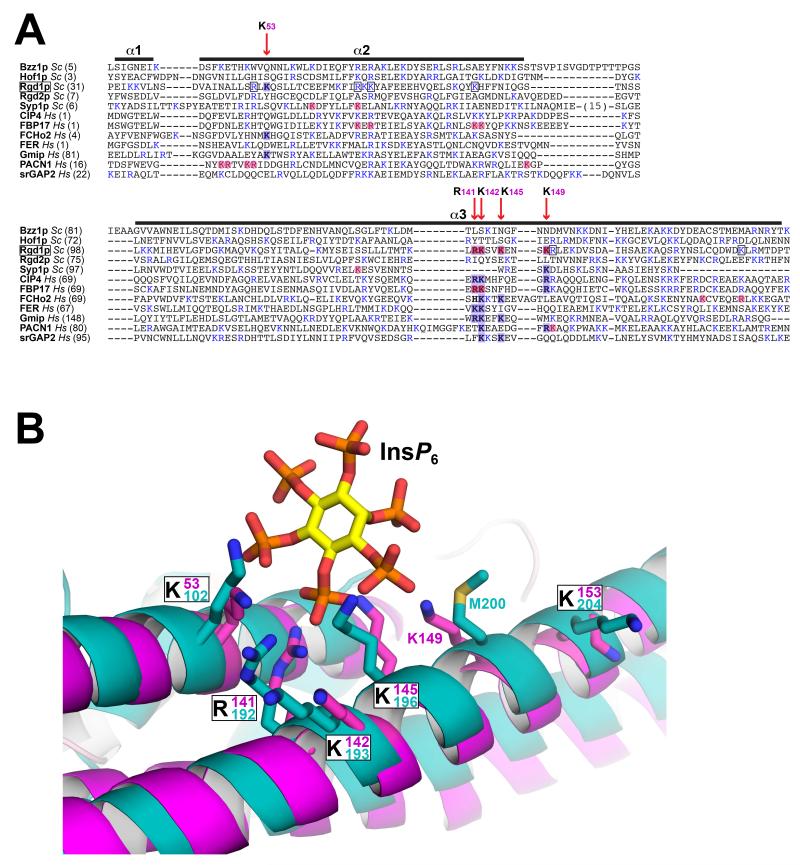
Figure 6. Conservation of InsP6 binding site elements in other F-BAR domains.
(A) The S. cerevisiae and human F-BAR domains listed were aligned using PROMALS3D (Pei and Grishin, 2014), which includes evolutionary and structural information. Only helices α1 to α3 are shown, corresponding to the first half of the F-BAR domain, which contains the Rgd1p InsP6 binding site. Basic residues are colored blue. Positions of key residues in the Rgd1p F-BAR InsP6 binding site are depicted with red vertical arrows, labeled with the Rgd1p residue number – and shaded blue where they occur in Rgd1p or other F-BAR domains. Residues shaded light red correspond to residues implicated in binding of other F-BAR domains to anionic membranes by mutational studies. Where a basic residue is also present at one of these positions in Rgd1p, it is boxed. (B) Close-up view of the Rgd1p F-BAR (magenta) InsP6 binding site overlaid with the structure of the Gmip (GEM interacting protein – a Rho-GAP) F-BAR domain shown in cyan (PDB entry 3QWE). The two F-BAR domain structures are compared in Figure S5. The InsP6 binding site is almost completely conserved in Gmip F-BAR, with the exception of K149 (replaced by M200, but with K201 close by).