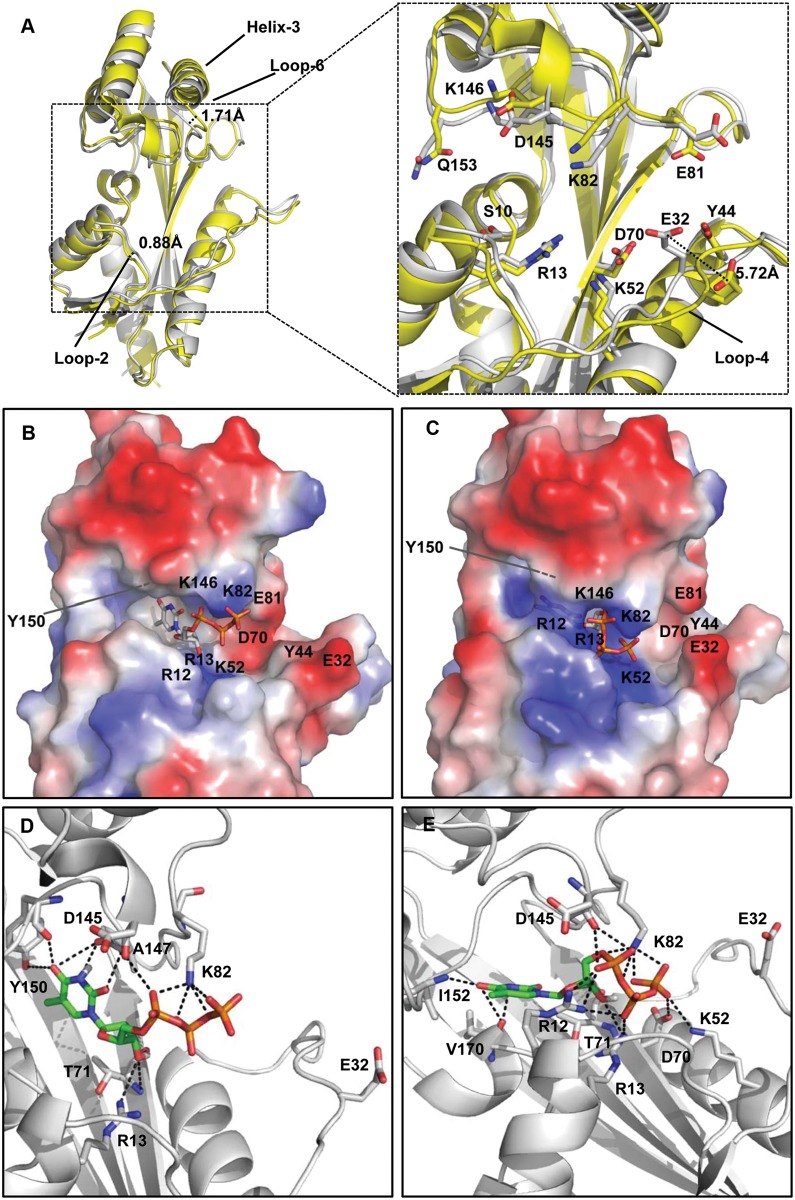
Fig 4. Structure details.
(A) The overall aligned structure of YhdE_E33A and detailed structure of substrate-binding pocket in YhdE_E33A open conformation (yellow, PDB code 4P0U) and closed conformation (white, PDB code 4P0E). Residues involved in substrate binding or implicated in the reaction are shown as sticks. Conformational changes are presented based on the distance between the two C-α atoms on the same residue of main chain in loop-6 near helix-3 (1.71 Å) and loop-2 (0.88 Å). The conformational shift of the E32 residue between the open and closed conformations is shown in the detailed figure on the right. Distance measurement was performed by aligning the two conformations and measuring the distance between the two CD atoms in the carboxyl group of the two E32 residues. Comparison of YhdE surface charge in the open (B) and closed conformations (C). Blue and red indicate positive and negative charge distribution on the YhdE surface, respectively. Binding model of dTTP in the YhdE open (D) and closed conformations (E). The amino acids involved in the interaction with dTTP are presented with all carbon atoms colored white. dTTP is presented as all carbon atoms colored green. E32 is also shown in each figure.