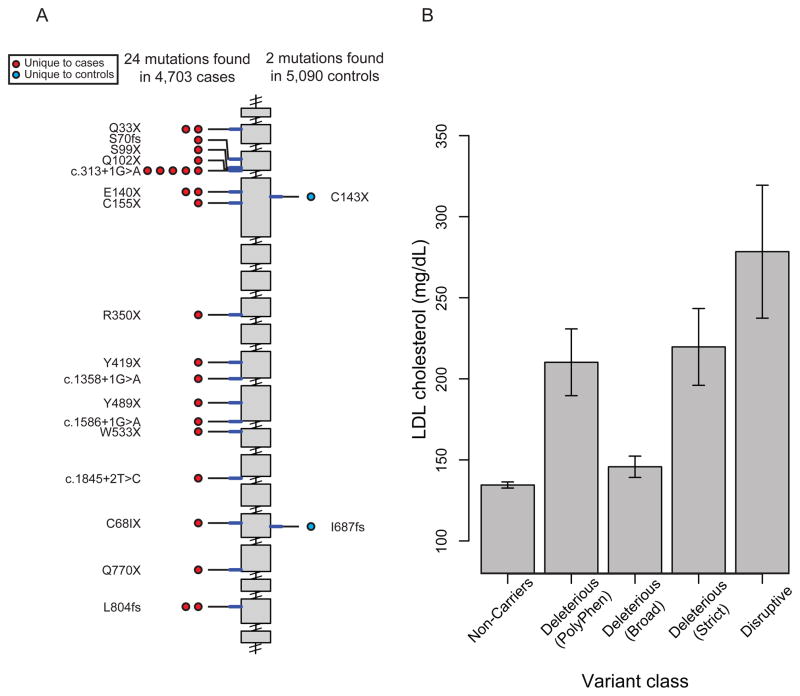
Figure 3. Low-density lipoprotein receptor (LDLR) mutations discovered after sequencing 9,793 individuals.
A. Individual disruptive mutations (nonsense, indel frameshift, and splice-site variants with minor allele frequency less than 1%) are depicted according to genomic position along the length of the LDLR gene starting at the 5′ end (top). The number of circles on the left and right represents the number of times that mutation is observed in cases or controls, respectively. Mutations are shaded in red or blue, if observed in cases only or controls only, respectively. B. Low-density lipoprotein cholesterol level as a function of LDLR gene mutation annotation. Mean (height of bar) and 95 % confidence intervals (error bars) are shown. Each individual is categorized based on mutation annotation as follows. Non-Carriers: carriers without a missense or disruptive mutation; “Deleterious (PolyPhen)” as defined by nonsense, splice-site, indel frameshift, and missense annotated as “possibly damaging” or “probably damaging” by PolyPhen-2 HumDiv software; “Deleterious (Broad)” as defined by nonsense, splice-site, indel frameshift, and missense annotated as deleterious by at least one of five protein prediction algorithms (LRT score, MutationTaster, PolyPhen-2 HumDiv, PolyPhen-2 HumVar and SIFT); “Deleterious (Strict)” as defined by nonsense, splice-site, indel frameshift, and missense annotated as deleterious by all five of the above protein prediction algorithms; Disruptive: carriers of mutations that are nonsense, indel frameshift, or splice-site.