Figure 2.
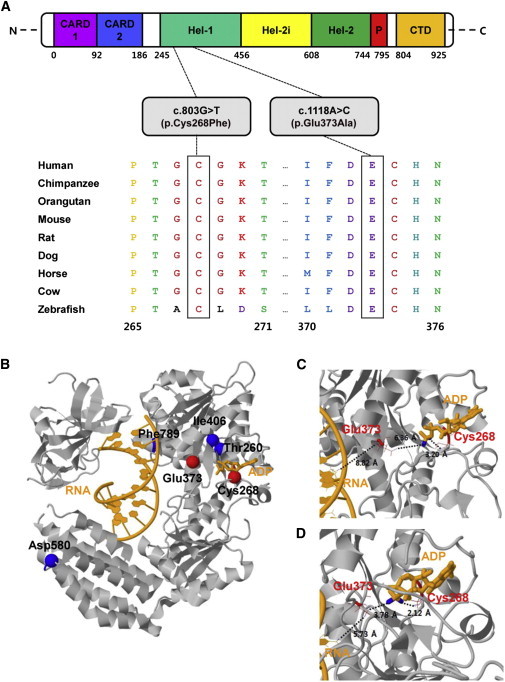
Ortholog Conservation of DDX58 Mutations and Molecular Helicase Structure of DDX58
(A) Schematic representation of DDX58 mutations relative to the affected protein domains. Also, sequence alignment of DDX58 is shown in vertebrate species. The alignment regions corresponding to the missense mutations are shown. Cys at codon 268 and Glu at codon 373 are highly conserved across all species. The Cys and Glu residues are indicated by the closed white boxes. The Ensembl IDs for DDX58 sequences aligned in this study are as follows: human, ENSP00000369197; chimpanzee, ENSPTRT00000038562; orangutan, ENSPPYT00000022324; mouse, ENSMUSP00000115052; rat, ENSRNOT00000008465; dog, ENSCAFT00000002841; horse, ENSECAT00000023725; cow, ENSBTAP00000053514; and zebrafish, ENSDART00000058276. Abbreviations are as follows: CARD, caspase activation recruitment domain; CTD, C-terminal domain; Hel, helicase domain (Hel-1 and Hel-2 are the two conserved core helicase domains, and Hel-2i is an insertion domain conserved in DDX58-like helicase family); and P, pincer of bridge region connecting Hel-2 to the CTD involved in binding dsRNA.
(B) The protein structure is drawn in gray. The RNA and ADP molecules are shown in orange. The locations of the amino acid changes caused by four known SNPs (rs35527044 [p.Thr260Pro], rs951618 [p.Ile406Thr], rs17217280 [p.Asp580Glu], and rs35253851 [p.Phe789Leu]) and two most likely pathogenic variants (p.Cys268Phe and p.Glu373Ala) are shown by blue and red spheres, respectively. The Thr260, Ile406, and Phe789 sites are hidden by the protein. PDB ID 3ZD7 was used for the wild-type (WT) structure of DDX58.8 This structure contains 73 experimentally missing residues. All protein structural analyses were executed with the program CHARMM.9
(C and D) Focused views of the WT X-ray structure (C) and 10-ns (ns) molecular dynamics (MD) final structure (D). The 10-ns MD simulation was performed with an implicit solvation model, i.e., the generalized Born model with a simple switch.10 Two residues (Cys268 and Glu373) affected by mutations are drawn as red wireframe models. Close distances are shown as black dotted lines with distance values. The used atom pairs for the distance measurements are shown in Table S3.
